TANGO1 recruits Sec16 to coordinately organize ER exit sites for efficient secretion
- PMID: 28442536
- PMCID: PMC5461033
- DOI: 10.1083/jcb.201703084
TANGO1 recruits Sec16 to coordinately organize ER exit sites for efficient secretion
Abstract
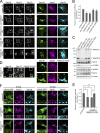
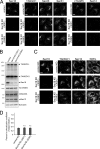
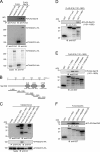
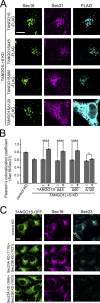
Mammalian endoplasmic reticulum (ER) exit sites export a variety of cargo molecules including oversized cargoes such as collagens. However, the mechanisms of their assembly and organization are not fully understood. TANGO1L is characterized as a collagen receptor, but the function of TANGO1S remains to be investigated. Here, we show that direct interaction between both isoforms of TANGO1 and Sec16 is not only important for their correct localization but also critical for the organization of ER exit sites. The depletion of TANGO1 disassembles COPII components as well as membrane-bound ER-resident complexes, resulting in fewer functional ER exit sites and delayed secretion. The ectopically expressed TANGO1 C-terminal domain responsible for Sec16 binding in mitochondria is capable of recruiting Sec16 and other COPII components. Moreover, TANGO1 recruits membrane-bound macromolecular complexes consisting of cTAGE5 and Sec12 to the ER exit sites. These data suggest that mammalian ER exit sites are organized by TANGO1 acting as a scaffold, in cooperation with Sec16 for efficient secretion.
© 2017 Maeda et al.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

