Zinc-Dependent Transcriptional Regulation in Paracoccus denitrificans
- PMID: 28443074
- PMCID: PMC5387054
- DOI: 10.3389/fmicb.2017.00569
Zinc-Dependent Transcriptional Regulation in Paracoccus denitrificans
Abstract
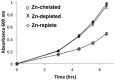
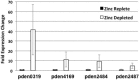
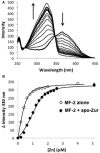
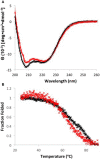
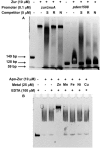
Zinc homeostasis is critical for bacterial survival and is mediated largely at the transcriptional level by the regulation of zinc uptake and efflux genes. Here we use RNA-seq to assess transcriptional changes as a result of zinc limitation in the denitrifying bacterium Paracoccus denitrificans. The results identify the differential expression of 147 genes, most of which were upregulated in zinc-depleted medium. Included in this set of genes are a large number of transition metal transporters, several transcription factors, and hypothetical proteins. Intriguingly, genes encoding nitric oxide reductase (norCB) and nitrite reductase (nirS) were also upregulated. A Zur consensus binding motif was identified in the promoters of the most highly upregulated genes. The zinc uptake regulator (Zur) from this organism was also characterized and shown to bind to the Zur motif in a zinc-dependent manner. This work expands our current understanding of the transcriptional response of gram-negative bacteria to zinc limitation and identifies genes involved in denitrification as part of the Zur regulon.
Keywords: RNA-seq; gene regulation; metal homeostasis; transcription factors; zinc.
Figures
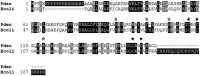


References
-
- Anderegg G. H. E., Podder N. G., Wenk F. (1977). Pyridinderivate als Komplexbildner. XI1. Die thermodynamik der metallkomplexbildung mit Bis-, Tris-, and Tetrakis[(2-pyridyl)methyl]-aminen. Helv. Chem. Acta 60, 123–140. 10.1002/hlca.19770600115 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

