Genetic and Dietary Iron Overload Differentially Affect the Course of Salmonella Typhimurium Infection
- PMID: 28443246
- PMCID: PMC5387078
- DOI: 10.3389/fcimb.2017.00110
Genetic and Dietary Iron Overload Differentially Affect the Course of Salmonella Typhimurium Infection
Abstract
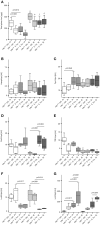
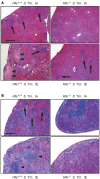
Genetic and dietary forms of iron overload have distinctive clinical and pathophysiological features. HFE-associated hereditary hemochromatosis is characterized by overwhelming intestinal iron absorption, parenchymal iron deposition, and macrophage iron depletion. In contrast, excessive dietary iron intake results in iron deposition in macrophages. However, the functional consequences of genetic and dietary iron overload for the control of microbes are incompletely understood. Using Hfe+/+ and Hfe-/- mice in combination with oral iron overload in a model of Salmonella enterica serovar Typhimurium infection, we found animals of either genotype to induce hepcidin antimicrobial peptide expression and hypoferremia following systemic infection in an Hfe-independent manner. As predicted, Hfe-/- mice, a model of hereditary hemochromatosis, displayed reduced spleen iron content, which translated into improved control of Salmonella replication. Salmonella adapted to the iron-poor microenvironment in the spleens of Hfe-/- mice by inducing the expression of its siderophore iron-uptake machinery. Dietary iron loading resulted in higher bacterial numbers in both WT and Hfe-/- mice, although Hfe deficiency still resulted in better pathogen control and improved survival. This suggests that Hfe deficiency may exert protective effects in addition to the control of iron availability for intracellular bacteria. Our data show that a dynamic adaptation of iron metabolism in both immune cells and microbes shapes the host-pathogen interaction in the setting of systemic Salmonella infection. Moreover, Hfe-associated iron overload and dietary iron excess result in different outcomes in infection, indicating that tissue and cellular iron distribution determines the susceptibility to infection with specific pathogens.
Keywords: Salmonella; hepcidin; infection; iron; lipocalin; macrophage; siderophore.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

