LigParGen web server: an automatic OPLS-AA parameter generator for organic ligands
- PMID: 28444340
- PMCID: PMC5793816
- DOI: 10.1093/nar/gkx312
LigParGen web server: an automatic OPLS-AA parameter generator for organic ligands
Abstract
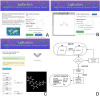
The accurate calculation of protein/nucleic acid-ligand interactions or condensed phase properties by force field-based methods require a precise description of the energetics of intermolecular interactions. Despite the progress made in force fields, small molecule parameterization remains an open problem due to the magnitude of the chemical space; the most critical issue is the estimation of a balanced set of atomic charges with the ability to reproduce experimental properties. The LigParGen web server provides an intuitive interface for generating OPLS-AA/1.14*CM1A(-LBCC) force field parameters for organic ligands, in the formats of commonly used molecular dynamics and Monte Carlo simulation packages. This server has high value for researchers interested in studying any phenomena based on intermolecular interactions with ligands via molecular mechanics simulations. It is free and open to all at jorgensenresearch.com/ligpargen, and has no login requirements.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Sponer J., Banas P., Jurecka P., Zgarbova M., Kuhrova P., Havrila M., Krepl M., Stadlbauer P., Otyepka M.. Molecular dynamics simulations of nucleic acids. From tetranucleotides to the ribosome. J. Phys. Chem. Lett. 2014; 5:1771–1782. - PubMed
-
- Best R.B., Zhu, Shim X., Lopes J., Mittal P.E., Feig J., Mackerell M., MacKerell A.D. Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi (1) and chi (2) dihedral angles. J. Chem. Theory Comput. 2012; 8:3257–3273. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

