Polyglutamine tracts regulate beclin 1-dependent autophagy
- PMID: 28445460
- PMCID: PMC5420314
- DOI: 10.1038/nature22078
Polyglutamine tracts regulate beclin 1-dependent autophagy
Abstract
Nine neurodegenerative diseases are caused by expanded polyglutamine (polyQ) tracts in different proteins, such as huntingtin in Huntington's disease and ataxin 3 in spinocerebellar ataxia type 3 (SCA3). Age at onset of disease decreases with increasing polyglutamine length in these proteins and the normal length also varies. PolyQ expansions drive pathogenesis in these diseases, as isolated polyQ tracts are toxic, and an N-terminal huntingtin fragment comprising exon 1, which occurs in vivo as a result of alternative splicing, causes toxicity. Although such mutant proteins are prone to aggregation, toxicity is also associated with soluble forms of the proteins. The function of the polyQ tracts in many normal cytoplasmic proteins is unclear. One such protein is the deubiquitinating enzyme ataxin 3 (refs 7, 8), which is widely expressed in the brain. Here we show that the polyQ domain enables wild-type ataxin 3 to interact with beclin 1, a key initiator of autophagy. This interaction allows the deubiquitinase activity of ataxin 3 to protect beclin 1 from proteasome-mediated degradation and thereby enables autophagy. Starvation-induced autophagy, which is regulated by beclin 1, was particularly inhibited in ataxin-3-depleted human cell lines and mouse primary neurons, and in vivo in mice. This activity of ataxin 3 and its polyQ-mediated interaction with beclin 1 was competed for by other soluble proteins with polyQ tracts in a length-dependent fashion. This competition resulted in impairment of starvation-induced autophagy in cells expressing mutant huntingtin exon 1, and this impairment was recapitulated in the brains of a mouse model of Huntington's disease and in cells from patients. A similar phenomenon was also seen with other polyQ disease proteins, including mutant ataxin 3 itself. Our data thus describe a specific function for a wild-type polyQ tract that is abrogated by a competing longer polyQ mutation in a disease protein, and identify a deleterious function of such mutations distinct from their propensity to aggregate.
Conflict of interest statement
Potential competing financial interests: F.M.M. is currently an employee of Eli Lilly & Co. Ltd.
Figures
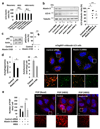
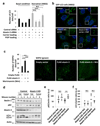


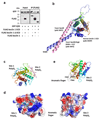
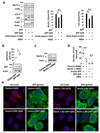
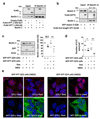
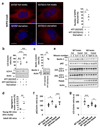
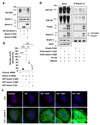



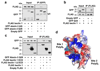

Comment in
-
Neurodegeneration: Role of repeats in protein clearance.Nature. 2017 May 4;545(7652):33-34. doi: 10.1038/nature22489. Epub 2017 Apr 26. Nature. 2017. PMID: 28445458 No abstract available.
-
Mechanisms of diseases: Excessive polyQ tracts curb autophagy.Nat Rev Mol Cell Biol. 2017 Jun;18(6):344. doi: 10.1038/nrm.2017.50. Epub 2017 May 10. Nat Rev Mol Cell Biol. 2017. PMID: 28488702 No abstract available.
-
Long polyglutamine tracts impair the autophagy protein quality control pathway.Mov Disord. 2017 Oct;32(10):1376. doi: 10.1002/mds.27118. Epub 2017 Aug 7. Mov Disord. 2017. PMID: 28782847 No abstract available.
References
-
- DiFiglia M, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. - PubMed
-
- Riess O, Rub U, Pastore A, Bauer P, Schols L. SCA3: neurological features, pathogenesis and animal models. Cerebellum. 2008;7:125–137. - PubMed
-
- Imarisio S, et al. Huntington's disease: from pathology and genetics to potential therapies. The Biochemical journal. 2008;412:191–209. - PubMed
-
- Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

