Various mutations compensate for a deleterious lacZα insert in the replication enhancer of M13 bacteriophage
- PMID: 28445507
- PMCID: PMC5405960
- DOI: 10.1371/journal.pone.0176421
Various mutations compensate for a deleterious lacZα insert in the replication enhancer of M13 bacteriophage
Abstract
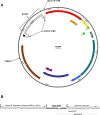
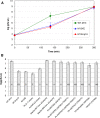
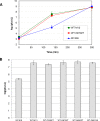
M13 and other members of the Ff class of filamentous bacteriophages have been extensively employed in myriad applications. The Ph.D. series of phage-displayed peptide libraries were constructed from the M13-based vector M13KE. As a direct descendent of M13mp19, M13KE contains the lacZα insert in the intergenic region between genes IV and II, where it interrupts the replication enhancer of the (+) strand origin. Phage carrying this 816-nucleotide insert are viable, but propagate in E. coli at a reduced rate compared to wild-type M13 phage, presumably due to a replication defect caused by the insert. We have previously reported thirteen compensatory mutations in the 5'-untranslated region of gene II, which encodes the replication initiator protein gIIp. Here we report several additional mutations in M13KE that restore a wild-type propagation rate. Several clones from constrained-loop variable peptide libraries were found to have ejected the majority of lacZα gene in order to reconstruct the replication enhancer, albeit with a small scar. In addition, new point mutations in the gene II 5'-untranslated region or the gene IV coding sequence have been spontaneously observed or synthetically engineered. Through phage propagation assays, we demonstrate that all these genetic modifications compensate for the replication defect in M13KE and restore the wild-type propagation rate. We discuss the mechanisms by which the insertion and ejection of the lacZα gene, as well as the mutations in the regulatory region of gene II, influence the efficiency of replication initiation at the (+) strand origin. We also examine the presence and relevance of fast-propagating mutants in phage-displayed peptide libraries.
Conflict of interest statement
Figures
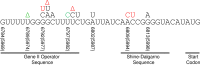
References
-
- Cao B, Yang M, Mao C. Phage as a Genetically Modifiable Supramacromolecule in Chemistry, Materials and Medicine. Acc Chem Res 2016. June 21;49(6):1111–1120. doi: 10.1021/acs.accounts.5b00557 - DOI - PMC - PubMed
-
- Henry KA, Arbabi-Ghahroudi M, Scott JK. Beyond phage display: non-traditional applications of the filamentous bacteriophage as a vaccine carrier, therapeutic biologic, and bioconjugation scaffold. Front Microbiol 2015. August 4;6:755 doi: 10.3389/fmicb.2015.00755 - DOI - PMC - PubMed
-
- Hamzeh-Mivehroud M, Alizadeh AA, Morris MB, Church WB, Dastmalchi S. Phage display as a technology delivering on the promise of peptide drug discovery. Drug Discov Today 2013. December;18(23–24):1144–1157. doi: 10.1016/j.drudis.2013.09.001 - DOI - PubMed
-
- Messing J. Phage M13 for the treatment of Alzheimer and Parkinson disease. Gene 2016. June 1;583(2):85–89. doi: 10.1016/j.gene.2016.02.005 - DOI - PubMed
-
- Sattar S, Bennett NJ, Wen WX, Guthrie JM, Blackwell LF, Conway JF, et al. Ff-nano, short functionalized nanorods derived from Ff (f1, fd, or M13) filamentous bacteriophage. Front Microbiol 2015. April 20;6:316 doi: 10.3389/fmicb.2015.00316 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

