Comparative genomic analysis between Corynebacterium pseudotuberculosis strains isolated from buffalo
- PMID: 28445543
- PMCID: PMC5406005
- DOI: 10.1371/journal.pone.0176347
Comparative genomic analysis between Corynebacterium pseudotuberculosis strains isolated from buffalo
Abstract
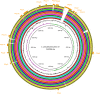
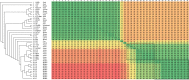
Corynebacterium pseudotuberculosis is a Gram-positive, pleomorphic, facultative intracellular pathogen that causes Oedematous Skin Disease (OSD) in buffalo. To better understand the pathogenic mechanisms of OSD, we performed a comparative genomic analysis of 11 strains of C. pseudotuberculosis isolated from different buffalo found to be infected in Egypt during an outbreak that occurred in 2008. Sixteen previously described pathogenicity islands (PiCp) were present in all of the new buffalo strains, but one of them, PiCp12, had an insertion that contained both a corynephage and a diphtheria toxin gene, both of which may play a role in the adaptation of C. pseudotuberculosis to this new host. Synteny analysis showed variations in the site of insertion of the corynephage during the same outbreak. A gene functional comparison showed the presence of a nitrate reductase operon that included genes involved in molybdenum cofactor biosynthesis, which is necessary for a positive nitrate reductase phenotype and is a possible adaptation for intracellular survival. Genomes from the buffalo strains also had fusions in minor pilin genes in the spaA and spaD gene cluster (spaCX and spaYEF), which could suggest either an adaptation to this particular host, or mutation events in the immediate ancestor before this particular epidemic. A phylogenomic analysis confirmed a clear separation between the Ovis and Equi biovars, but also showed what appears to be a clustering by host species within the Equi strains.
Conflict of interest statement
Figures





Similar articles
-
Oedematous skin disease of buffalo in Egypt.J Vet Med B Infect Dis Vet Public Health. 2001 May;48(4):241-58. doi: 10.1046/j.1439-0450.2001.00451.x. J Vet Med B Infect Dis Vet Public Health. 2001. PMID: 15129580 Review.
-
Assessing the Genotypic Differences between Strains of Corynebacterium pseudotuberculosis biovar equi through Comparative Genomics.PLoS One. 2017 Jan 26;12(1):e0170676. doi: 10.1371/journal.pone.0170676. eCollection 2017. PLoS One. 2017. PMID: 28125655 Free PMC article.
-
Corynebacterium pseudotuberculosis may be under anagenesis and biovar Equi forms biovar Ovis: a phylogenic inference from sequence and structural analysis.BMC Microbiol. 2016 Jun 2;16:100. doi: 10.1186/s12866-016-0717-4. BMC Microbiol. 2016. PMID: 27251711 Free PMC article.
-
Molecular epidemiology of Corynebacterium pseudotuberculosis isolated from horses in California.Infect Genet Evol. 2017 Apr;49:186-194. doi: 10.1016/j.meegid.2016.12.011. Epub 2016 Dec 13. Infect Genet Evol. 2017. PMID: 27979735
-
A description of genes of Corynebacterium pseudotuberculosis useful in diagnostics and vaccine applications.Genet Mol Res. 2008 Mar 18;7(1):252-60. doi: 10.4238/vol7-1gmr438. Genet Mol Res. 2008. PMID: 18551390 Review.
Cited by
-
Resequencing and characterization of the first Corynebacterium pseudotuberculosis genome isolated from camel.PeerJ. 2024 Jan 30;12:e16513. doi: 10.7717/peerj.16513. eCollection 2024. PeerJ. 2024. PMID: 38313017 Free PMC article.
-
CoryneRegNet 7, the reference database and analysis platform for corynebacterial gene regulatory networks.Sci Data. 2020 May 11;7(1):142. doi: 10.1038/s41597-020-0484-9. Sci Data. 2020. PMID: 32393779 Free PMC article.
-
Corynebacteria of the diphtheriae Species Complex in Companion Animals: Clinical and Microbiological Characterization of 64 Cases from France.Microbiol Spectr. 2023 Jun 15;11(3):e0000623. doi: 10.1128/spectrum.00006-23. Epub 2023 Apr 6. Microbiol Spectr. 2023. PMID: 37022195 Free PMC article.
-
First genome sequencing and comparative analyses of Corynebacterium pseudotuberculosis strains from Mexico.Stand Genomic Sci. 2018 Oct 10;13:21. doi: 10.1186/s40793-018-0325-z. eCollection 2018. Stand Genomic Sci. 2018. PMID: 30338024 Free PMC article.
-
Rapidly evolving changes and gene loss associated with host switching in Corynebacterium pseudotuberculosis.PLoS One. 2018 Nov 12;13(11):e0207304. doi: 10.1371/journal.pone.0207304. eCollection 2018. PLoS One. 2018. PMID: 30419061 Free PMC article.
References
-
- Dorella FA, Carvalho Pacheco L, Oliveira SC, Miyoshi A, Azevedo V. Corynebacterium pseudotuberculosis: microbiology, biochemical properties, pathogenesis and molecular studies of virulence. Vet Res. 2006;37: 201–218. doi: 10.1051/vetres:2005056 - DOI - PubMed
-
- Guaraldi AL de M, Júnior RH, Azevedo CVA de. Corynebacterium diphtheriae, Corynebacterium ulcerans and Corynebacterium pseudotuberculosis—General Aspects. In: Burkovski A, editor. Corynebacterium diphtheriae and Related Toxigenic Species. London, New York; 2014. pp. 15–37.
-
- Bernard K. The genus Corynebacterium and other medically relevant coryneform-like bacteria. J Clin Microbiol. 2012;50: 3152–8. doi: 10.1128/JCM.00796-12 - DOI - PMC - PubMed
-
- Oliveira A, Teixeira P, Azevedo M, Jamal SB, Tiwari S, Almeida S, et al. Corynebacterium pseudotuberculosis may be under anagenesis and biovar Equi forms biovar Ovis: a phylogenic inference from sequence and structural analysis. BMC Microbiol. BMC Microbiology; 2016;16: 100 doi: 10.1186/s12866-016-0717-4 - DOI - PMC - PubMed
-
- Biberstein EL, Knight HD, Jang S. Two biotypes of Corynebacterium pseudotuberculosis. Vet Rec. 1971;89: 691–692. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

