NMRbox: A Resource for Biomolecular NMR Computation
- PMID: 28445744
- PMCID: PMC5406371
- DOI: 10.1016/j.bpj.2017.03.011
NMRbox: A Resource for Biomolecular NMR Computation
Abstract
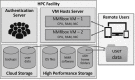
Advances in computation have been enabling many recent advances in biomolecular applications of NMR. Due to the wide diversity of applications of NMR, the number and variety of software packages for processing and analyzing NMR data is quite large, with labs relying on dozens, if not hundreds of software packages. Discovery, acquisition, installation, and maintenance of all these packages is a burdensome task. Because the majority of software packages originate in academic labs, persistence of the software is compromised when developers graduate, funding ceases, or investigators turn to other projects. To simplify access to and use of biomolecular NMR software, foster persistence, and enhance reproducibility of computational workflows, we have developed NMRbox, a shared resource for NMR software and computation. NMRbox employs virtualization to provide a comprehensive software environment preconfigured with hundreds of software packages, available as a downloadable virtual machine or as a Platform-as-a-Service supported by a dedicated compute cloud. Ongoing development includes a metadata harvester to regularize, annotate, and preserve workflows and facilitate and enhance data depositions to BioMagResBank, and tools for Bayesian inference to enhance the robustness and extensibility of computational analyses. In addition to facilitating use and preservation of the rich and dynamic software environment for biomolecular NMR, NMRbox fosters the development and deployment of a new class of metasoftware packages. NMRbox is freely available to not-for-profit users.
Copyright © 2017 Biophysical Society. All rights reserved.
Figures

Similar articles
-
Merging NMR Data and Computation Facilitates Data-Centered Research.Front Mol Biosci. 2022 Jan 17;8:817175. doi: 10.3389/fmolb.2021.817175. eCollection 2021. Front Mol Biosci. 2022. PMID: 35111815 Free PMC article. Review.
-
BioMagResBank (BMRB) as a Resource for Structural Biology.Methods Mol Biol. 2020;2112:187-218. doi: 10.1007/978-1-0716-0270-6_14. Methods Mol Biol. 2020. PMID: 32006287 Free PMC article.
-
CONNJUR Workflow Builder: a software integration environment for spectral reconstruction.J Biomol NMR. 2015 Jul;62(3):313-26. doi: 10.1007/s10858-015-9946-3. Epub 2015 Jun 12. J Biomol NMR. 2015. PMID: 26066803 Free PMC article.
-
Cloud Computing with iPlant Atmosphere.Curr Protoc Bioinformatics. 2013 Oct 15;43:9.15.1-9.15.20. doi: 10.1002/0471250953.bi0915s43. Curr Protoc Bioinformatics. 2013. PMID: 26270172
-
Updates in metabolomics tools and resources: 2014-2015.Electrophoresis. 2016 Jan;37(1):86-110. doi: 10.1002/elps.201500417. Epub 2015 Nov 17. Electrophoresis. 2016. PMID: 26464019 Review.
Cited by
-
Structural dynamics of human deoxyuridine 5'-triphosphate nucleotidohydrolase (dUTPase).Sci Rep. 2024 Oct 30;14(1):26081. doi: 10.1038/s41598-024-76548-x. Sci Rep. 2024. PMID: 39477983 Free PMC article.
-
Pterostilbene-Isothiocyanate Inhibits Proliferation of Human MG-63 Osteosarcoma Cells via Abrogating β-Catenin/TCF-4 Interaction-A Mechanistic Insight.ACS Omega. 2023 Nov 6;8(46):43474-43489. doi: 10.1021/acsomega.3c02732. eCollection 2023 Nov 21. ACS Omega. 2023. PMID: 38027335 Free PMC article.
-
Backbone and ILV side-chain NMR resonance assignments of the catalytic domain of human deubiquitinating enzyme USP7.Biomol NMR Assign. 2022 Oct;16(2):197-203. doi: 10.1007/s12104-022-10079-2. Epub 2022 May 10. Biomol NMR Assign. 2022. PMID: 35536398 Free PMC article.
-
ILV methyl NMR resonance assignments of the 81 kDa E. coli β-clamp.Biomol NMR Assign. 2022 Oct;16(2):317-323. doi: 10.1007/s12104-022-10097-0. Epub 2022 Jun 10. Biomol NMR Assign. 2022. PMID: 35687262 Free PMC article.
-
Reaching the sparse-sampling limit for reconstructing a single peak in a 2D NMR spectrum using iterated maps.J Biomol NMR. 2019 Nov;73(10-11):545-560. doi: 10.1007/s10858-019-00262-4. Epub 2019 Jul 10. J Biomol NMR. 2019. PMID: 31292847 Free PMC article.
References
-
- Ernst R.R. Without computers—no modern NMR. In: Hoch J.C., Poulsen F.M., Redfield C., editors. Computational Aspects of the Study of Biological Macromolecules by Nuclear Magnetic Resonance Spectroscopy. Plenum Press; New York: 1991. pp. 1–25.
-
- Hoch J.C. Modern spectrum analysis in nuclear magnetic resonance: alternatives to the Fourier transform. Methods Enzymol. 1989;176:216–241. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

