Physical and Functional Characterization of a Viral Genome Maturation Complex
- PMID: 28445747
- PMCID: PMC5406279
- DOI: 10.1016/j.bpj.2017.02.041
Physical and Functional Characterization of a Viral Genome Maturation Complex
Abstract
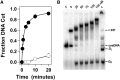
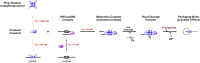
Genome packaging is strongly conserved in the complex double-stranded DNA viruses, including the herpesviruses and many bacteriophages. In these cases, viral DNA is packaged into a procapsid shell by a terminase enzyme. The packaging substrate is typically a concatemer composed of multiple genomes linked in a head-to-tail fashion, and terminase enzymes perform two essential functions: 1) excision of a unit length genome from the concatemer (genome maturation) and 2) translocation of the duplex into a procapsid (genome packaging). While the packaging motors have been described in some detail, the maturation complexes remain ill characterized. Here we describe the assembly, physical characteristics, and catalytic activity of the λ-genome maturation complex. The λ-terminase protomer is composed of one large catalytic subunit tightly associated with two DNA recognition subunits. The isolated protomer binds DNA weakly and does not discriminate between nonspecific DNA and duplexes that contain the packaging initiation sequence, cos. The Escherichia coli integration host factor protein (IHF) is required for efficient λ-development in vivo and a specific IHF recognition sequence is found within cos. We show that IHF and the terminase protomer cooperatively assemble at the cos site and that the small terminase subunit plays the dominant role in complex assembly. Analytical ultracentrifugation analysis reveals that the maturation complex is composed of four protomers and one IHF heterodimer bound at the cos site. Tetramer assembly activates the cos-cleavage nuclease activity of the enzyme, which matures the genome end in preparation for packaging. The stoichiometry and catalytic activity of the complex is reminiscent of the type IIE and IIF restriction endonucleases and the two systems may share mechanistic features. This study, to our knowledge, provides our first detailed glimpse into the structural and functional features of a viral genome maturation complex, an essential intermediate in the development of complex dsDNA viruses.
Copyright © 2017 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
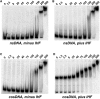
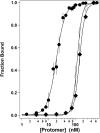
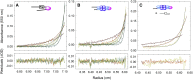
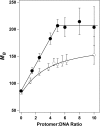
Similar articles
-
Bacteriophage lambda gpNu1 and Escherichia coli IHF proteins cooperatively bind and bend viral DNA: implications for the assembly of a genome-packaging motor.Biochemistry. 2006 Apr 25;45(16):5180-9. doi: 10.1021/bi052284b. Biochemistry. 2006. PMID: 16618107
-
Assembly of bacteriophage lambda terminase into a viral DNA maturation and packaging machine.Biochemistry. 2006 Dec 26;45(51):15259-68. doi: 10.1021/bi0615036. Epub 2006 Nov 30. Biochemistry. 2006. PMID: 17176048
-
Energy-independent helicase activity of a viral genome packaging motor.Biochemistry. 2012 Jan 10;51(1):391-400. doi: 10.1021/bi201604b. Epub 2011 Dec 30. Biochemistry. 2012. PMID: 22191393 Free PMC article.
-
Virus DNA packaging: the strategy used by phage lambda.Mol Microbiol. 1995 Jun;16(6):1075-86. doi: 10.1111/j.1365-2958.1995.tb02333.x. Mol Microbiol. 1995. PMID: 8577244 Review.
-
The terminase enzyme from bacteriophage lambda: a DNA-packaging machine.Cell Mol Life Sci. 2000 Jan 20;57(1):128-48. doi: 10.1007/s000180050503. Cell Mol Life Sci. 2000. PMID: 10949585 Free PMC article. Review.
Cited by
-
A novel Saclayvirus Acinetobacter baumannii phage genomic analysis and effectiveness in preventing pneumonia.Appl Microbiol Biotechnol. 2024 Jul 27;108(1):428. doi: 10.1007/s00253-024-13208-0. Appl Microbiol Biotechnol. 2024. PMID: 39066795 Free PMC article.
-
ATP serves as a nucleotide switch coupling the genome maturation and packaging motor complexes of a virus assembly machine.Nucleic Acids Res. 2020 May 21;48(9):5006-5015. doi: 10.1093/nar/gkaa205. Nucleic Acids Res. 2020. PMID: 32255177 Free PMC article.
-
Evidence that a catalytic glutamate and an 'Arginine Toggle' act in concert to mediate ATP hydrolysis and mechanochemical coupling in a viral DNA packaging motor.Nucleic Acids Res. 2019 Feb 20;47(3):1404-1415. doi: 10.1093/nar/gky1217. Nucleic Acids Res. 2019. PMID: 30541105 Free PMC article.
-
Functional Dissection of a Viral DNA Packaging Machine's Walker B Motif.J Mol Biol. 2019 Nov 8;431(22):4455-4474. doi: 10.1016/j.jmb.2019.08.012. Epub 2019 Aug 30. J Mol Biol. 2019. PMID: 31473160 Free PMC article.
-
Single-molecule measurements of bacteriophage lambda DNA packaging using purified terminase motor proteins and E. coli integration host factor.Sci Rep. 2025 Feb 27;15(1):7093. doi: 10.1038/s41598-024-74915-2. Sci Rep. 2025. PMID: 40016253 Free PMC article.
References
-
- Calendar R., Abedon S.T. Oxford University Press; New York, NY: 2006. The Bacteriophages.
-
- Roizman B., Knipe D.M., Whitley R.J. Herpes simplex viruses. In: Knipe D.M., Howley P.M., editors. Fields Virology. Lippincott Williams and Wilkins; New York, NY: 2007. pp. 2501–2602.
-
- Feiss M., Catalano C.E. Bacteriophage λ terminase and the mechanism of viral DNA packaging. In: Catalano C.E., editor. Viral Genome Packaging Machines: Genetics, Structure, and Mechanism. Kluwer Academic/Plenum Publishers; New York, NY: 2005. pp. 5–39.
-
- Baines J.D. Herpes simplex virus capsid assembly and DNA packaging: a present and future antiviral drug target. Trends Microbiol. 2011;19:606–613. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

