Signatures of co-evolutionary host-pathogen interactions in the genome of the entomopathogenic nematode Steinernema carpocapsae
- PMID: 28446150
- PMCID: PMC5405473
- DOI: 10.1186/s12862-017-0935-x
Signatures of co-evolutionary host-pathogen interactions in the genome of the entomopathogenic nematode Steinernema carpocapsae
Abstract
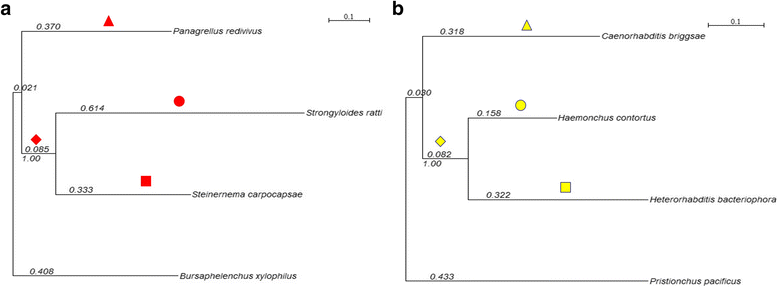
Background: The entomopathogenic nematode Steinernema carpocapsae has been used worldwide as a biocontrol agent for insect pests, making it an interesting model for understanding parasite-host interactions. Two models propose that these interactions are co-evolutionary processes in such a way that equilibrium is never reached. In one model, known as "arms race", new alleles in relevant genes are fixed in both host and pathogens by directional positive selection, producing recurrent and alternating selective sweeps. In the other model, known as"trench warfare", persistent dynamic fluctuations in allele frequencies are sustained by balancing selection. There are some examples of genes evolving according to both models, however, it is not clear to what extent these interactions might alter genome-level evolutionary patterns and intraspecific diversity. Here we investigate some of these aspects by studying genomic variation in S. carpocapsae and other pathogenic and free-living nematodes from phylogenetic clades IV and V.
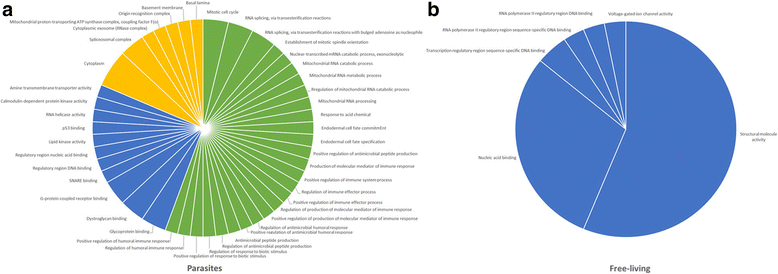
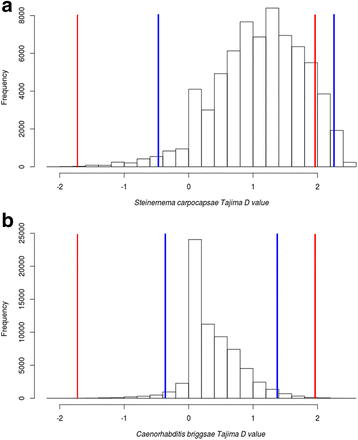
Results: To look for signatures of an arms-race dynamic, we conducted massive scans to detect directional positive selection in interspecific data. In free-living nematodes, we detected a significantly higher proportion of genes with sites under positive selection than in parasitic nematodes. However, in these genes, we found more enriched Gene Ontology terms in parasites. To detect possible effects of dynamic polymorphisms interactions we looked for signatures of balancing selection in intraspecific genomic data. The observed distribution of Tajima's D values in S. carpocapsae was more skewed to positive values and significantly different from the observed distribution in the free-living Caenorhabditis briggsae. Also, the proportion of significant positive values of Tajima's D was elevated in genes that were differentially expressed after induction with insect tissues as compared to both non-differentially expressed genes and the global scan.
Conclusions: Our study provides a first portrait of the effects that lifestyle might have in shaping the patterns of selection at the genomic level. An arms-race between hosts and pathogens seems to be affecting specific genetic functions but not necessarily increasing the number of positively selected genes. Trench warfare dynamics seem to be acting more generally in the genome, likely focusing on genes responding to the interaction, rather than targeting specific genetic functions.
Keywords: Arms race; Balancing selection; Genomic scans; Positive selection; Red Queen; Tajima’s D; Trench warfare; dN/dS.
Figures
References
-
- Platt HM. Foreword. In: Lorenzen S, editor. The Phylogenetic Systematics of Free-living Nematodes. London: The Ray Society; 1994. p. i–ii.
-
- Coghlan A. Nematode Genome Evolution. In: WormBook (ed. The C. elegans Research Community) 2005. http://www.wormbook.org. - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

