SCALE: modeling allele-specific gene expression by single-cell RNA sequencing
- PMID: 28446220
- PMCID: PMC5407026
- DOI: 10.1186/s13059-017-1200-8
SCALE: modeling allele-specific gene expression by single-cell RNA sequencing
Abstract
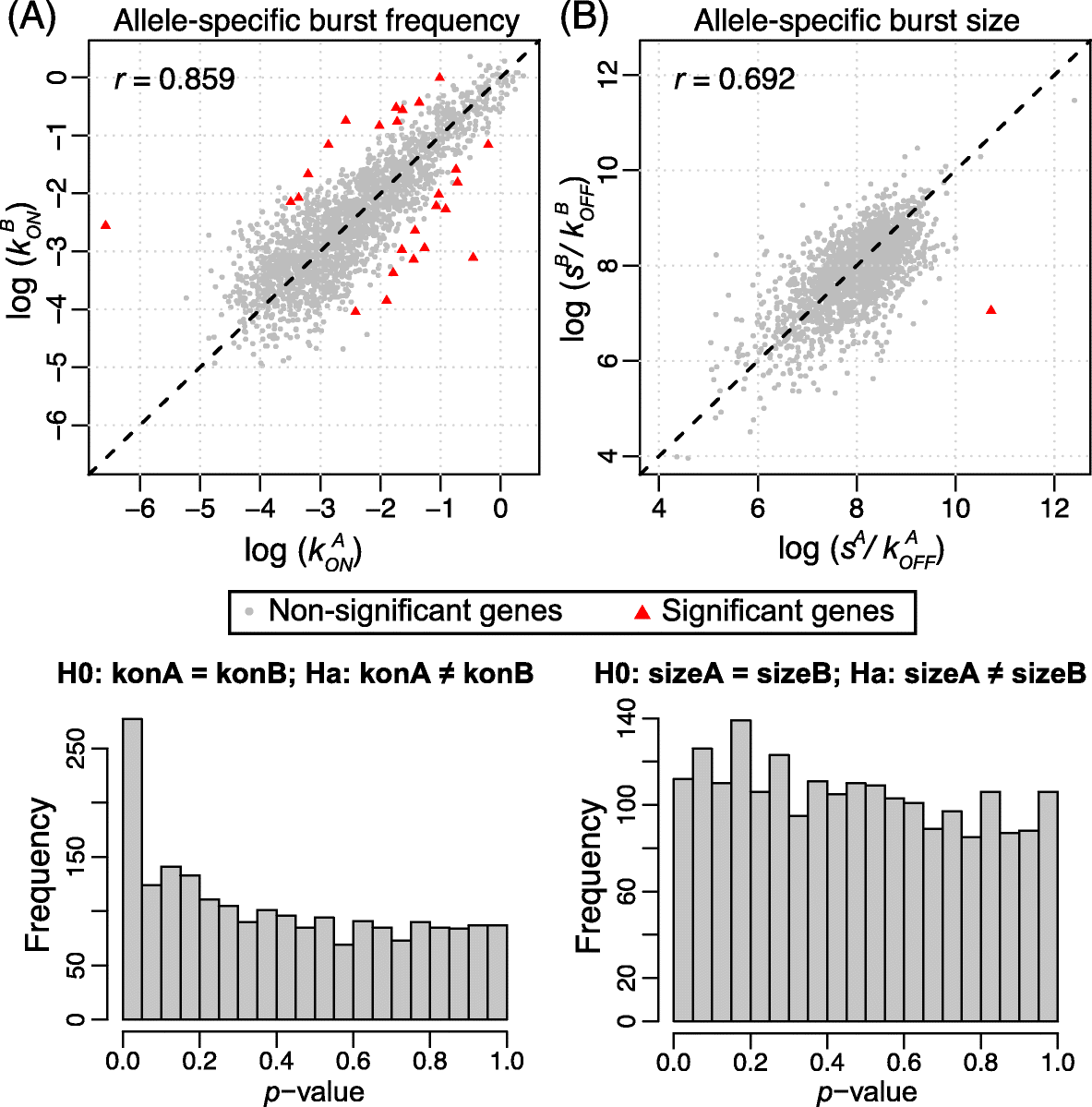
Allele-specific expression is traditionally studied by bulk RNA sequencing, which measures average expression across cells. Single-cell RNA sequencing allows the comparison of expression distribution between the two alleles of a diploid organism and the characterization of allele-specific bursting. Here, we propose SCALE to analyze genome-wide allele-specific bursting, with adjustment of technical variability. SCALE detects genes exhibiting allelic differences in bursting parameters and genes whose alleles burst non-independently. We apply SCALE to mouse blastocyst and human fibroblast cells and find that cis control in gene expression overwhelmingly manifests as differences in burst frequency.
Keywords: Allele-specific expression; Expression stochasticity; Single-cell RNA sequencing; Technical variability; Transcriptional bursting; cis and trans transcriptional control.
Figures
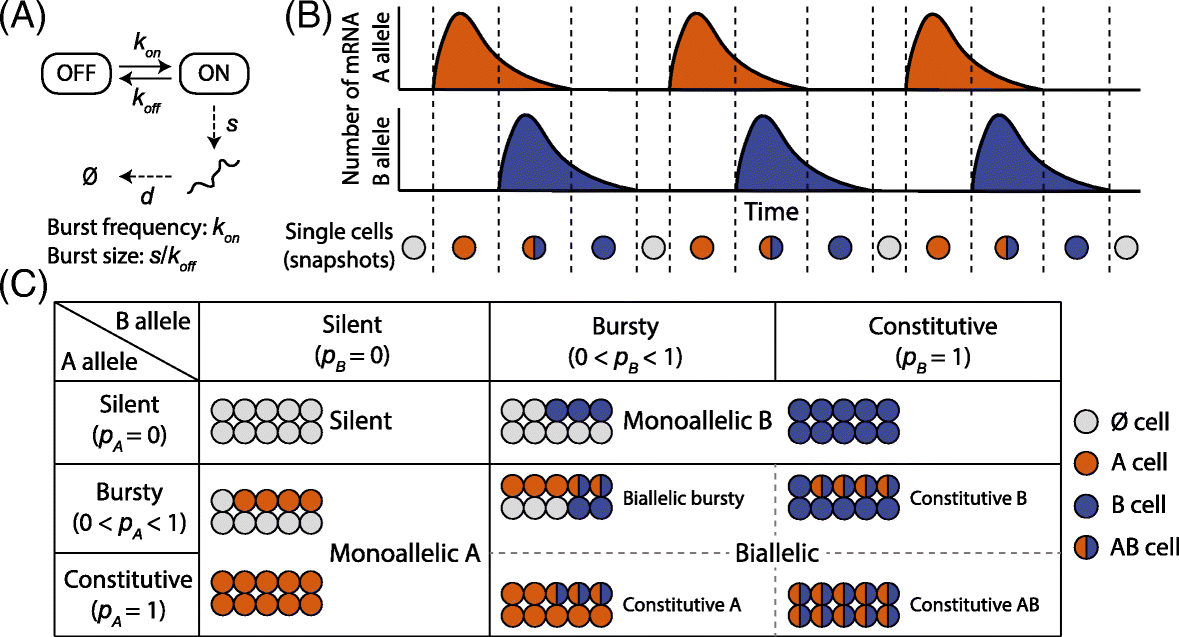
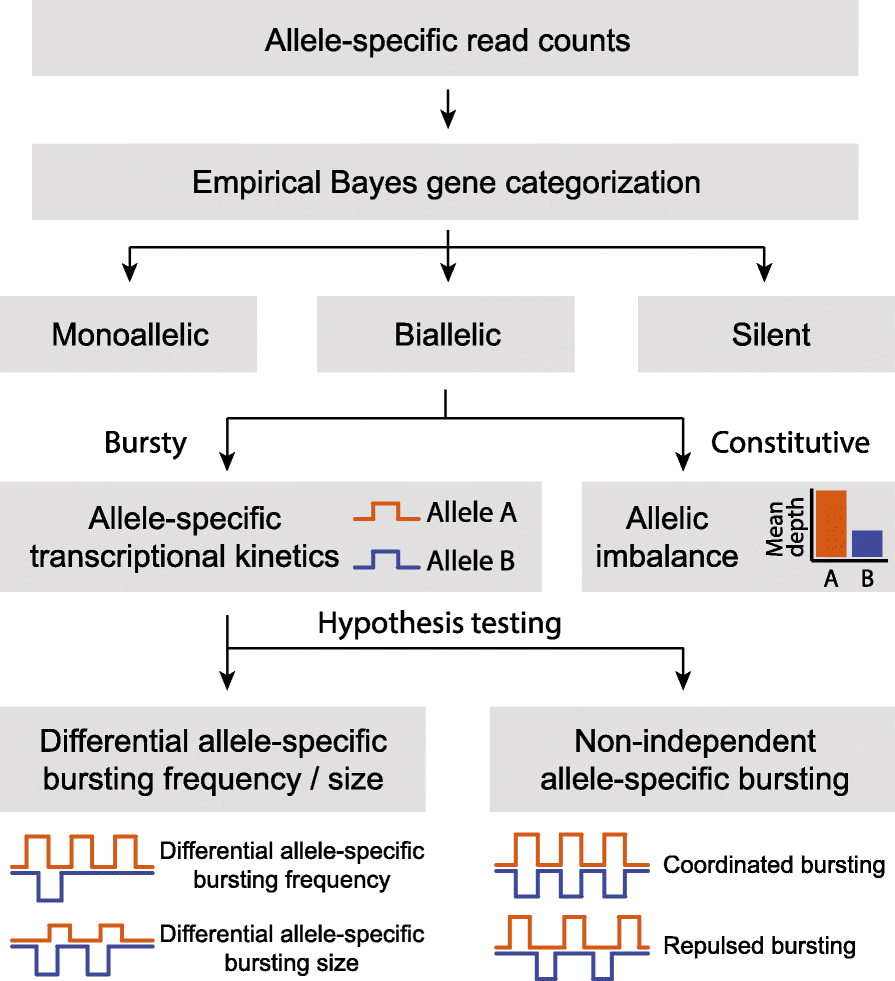

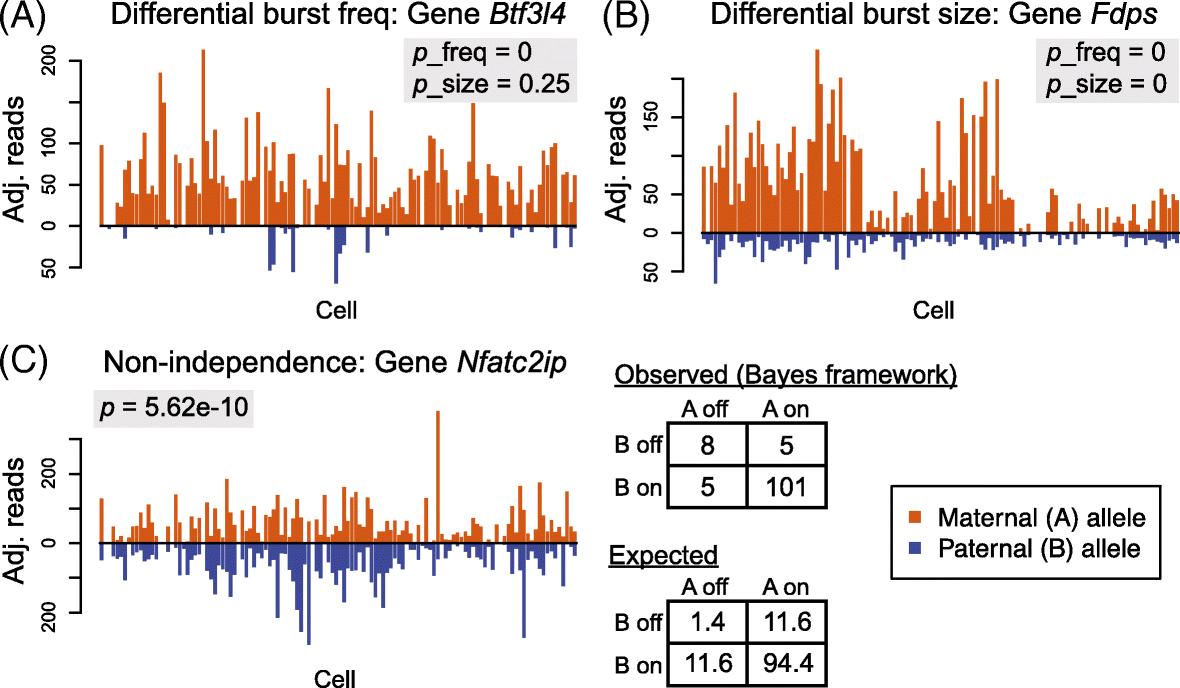
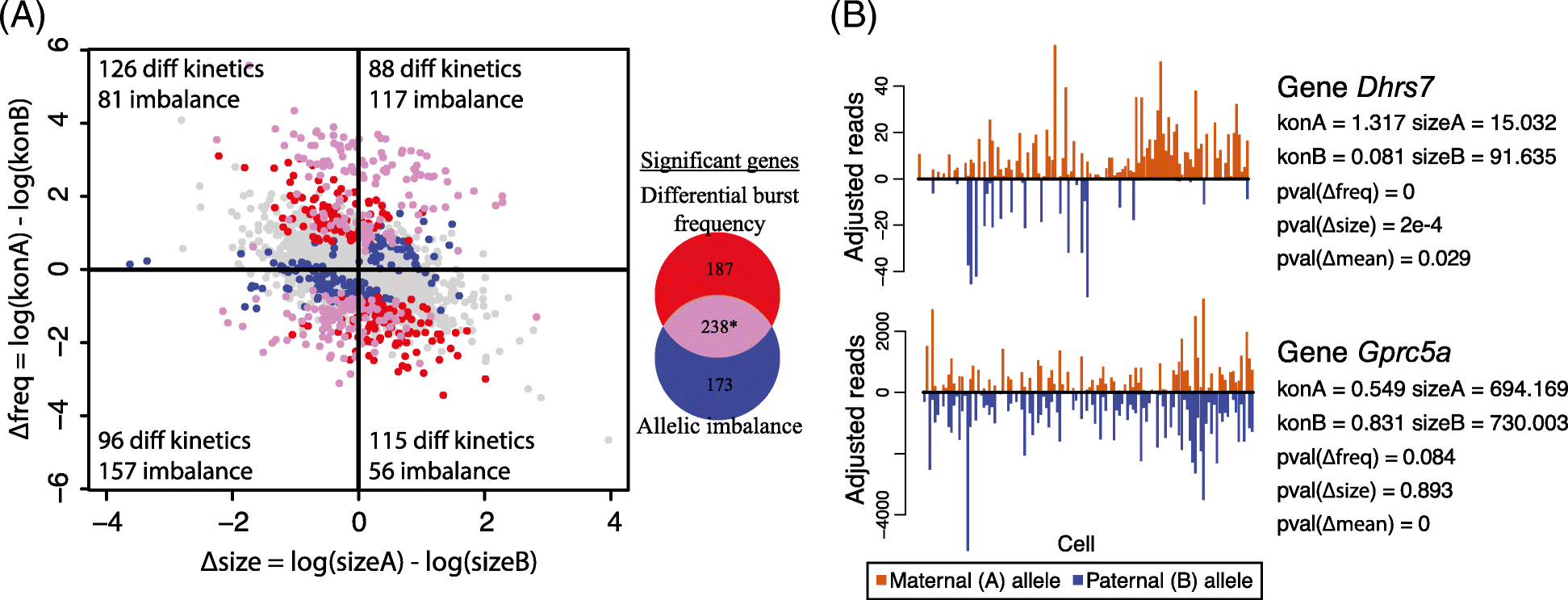
Similar articles
-
Single-Cell Allele-Specific Gene Expression Analysis.Methods Mol Biol. 2019;1935:155-174. doi: 10.1007/978-1-4939-9057-3_11. Methods Mol Biol. 2019. PMID: 30758826
-
Characterizing noise structure in single-cell RNA-seq distinguishes genuine from technical stochastic allelic expression.Nat Commun. 2015 Oct 22;6:8687. doi: 10.1038/ncomms9687. Nat Commun. 2015. PMID: 26489834 Free PMC article.
-
Single-cell RNA counting at allele and isoform resolution using Smart-seq3.Nat Biotechnol. 2020 Jun;38(6):708-714. doi: 10.1038/s41587-020-0497-0. Epub 2020 May 4. Nat Biotechnol. 2020. PMID: 32518404
-
The technology and biology of single-cell RNA sequencing.Mol Cell. 2015 May 21;58(4):610-20. doi: 10.1016/j.molcel.2015.04.005. Mol Cell. 2015. PMID: 26000846 Review.
-
Revealing allele-specific gene expression by single-cell transcriptomics.Int J Biochem Cell Biol. 2017 Sep;90:155-160. doi: 10.1016/j.biocel.2017.05.029. Epub 2017 May 31. Int J Biochem Cell Biol. 2017. PMID: 28578186 Review.
Cited by
-
Epigenomic states contribute to coordinated allelic transcriptional bursting in iPSC reprogramming.Life Sci Alliance. 2024 Feb 6;7(4):e202302337. doi: 10.26508/lsa.202302337. Print 2024 Apr. Life Sci Alliance. 2024. PMID: 38320809 Free PMC article.
-
BIRD: identifying cell doublets via biallelic expression from single cells.Bioinformatics. 2020 Jul 1;36(Suppl_1):i251-i257. doi: 10.1093/bioinformatics/btaa474. Bioinformatics. 2020. PMID: 32657402 Free PMC article.
-
STmut: a framework for visualizing somatic alterations in spatial transcriptomics data of cancer.Genome Biol. 2023 Nov 30;24(1):273. doi: 10.1186/s13059-023-03121-6. Genome Biol. 2023. PMID: 38037084 Free PMC article.
-
Canopy2: tumor phylogeny inference by bulk DNA and single-cell RNA sequencing.bioRxiv [Preprint]. 2024 Mar 19:2024.03.18.585595. doi: 10.1101/2024.03.18.585595. bioRxiv. 2024. PMID: 38562795 Free PMC article. Preprint.
-
Allele-specific DNA methylation and gene expression during shoot organogenesis in tissue culture of hybrid poplar.Hortic Res. 2024 Jan 24;11(3):uhae027. doi: 10.1093/hr/uhae027. eCollection 2024 Mar. Hortic Res. 2024. PMID: 38544548 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

