Dicer loss and recovery induce an oncogenic switch driven by transcriptional activation of the oncofetal Imp1-3 family
- PMID: 28446596
- PMCID: PMC5411708
- DOI: 10.1101/gad.296301.117
Dicer loss and recovery induce an oncogenic switch driven by transcriptional activation of the oncofetal Imp1-3 family
Erratum in
-
Corrigendum: Dicer loss and recovery induce an oncogenic switch driven by transcriptional activation of the oncofetal Imp1-3 family.Genes Dev. 2017 May 15;31(10):1066. doi: 10.1101/gad.302513.117. Genes Dev. 2017. PMID: 28637695 Free PMC article. No abstract available.
Abstract
MicroRNAs (miRNAs) are post-transcriptional regulators of gene expression critical for organismal viability. Changes in miRNA activity are common in cancer, but how these changes relate to subsequent alterations in transcription and the process of tumorigenesis is not well understood. Here, we report a deep transcriptional, oncogenic network regulated by miRNAs. We present analysis of the gene expression and phenotypic changes associated with global miRNA restoration in miRNA-deficient fibroblasts. This analysis uncovers a miRNA-repressed network containing oncofetal genes Imp1, Imp2, and Imp3 (Imp1-3) that is up-regulated primarily transcriptionally >100-fold upon Dicer loss and is resistant to resilencing by complete restoration of miRNA activity. This Dicer-resistant epigenetic switch confers tumorigenicity to these cells. Let-7 targets Imp1-3 are required for this tumorigenicity and feed back to reinforce and sustain expression of the oncogenic network. Together, these Dicer-resistant genes constitute an mRNA expression signature that is present in numerous human cancers and is associated with poor survival.
Keywords: Dicer; Imp1–3; cancer; let-7; miRNA; oncofetal.
© 2017 JnBaptiste et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
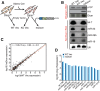
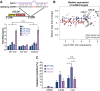
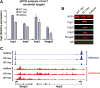

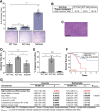

References
-
- Bahubeshi A, Bal N, Rio Frio T, Hamel N, Pouchet C, Yilmaz A, Bouron-Dal Soglio D, Williams GM, Tischkowitz M, Priest JR, et al. 2010. Germline DICER1 mutations and familial cystic nephroma. J Med Genet 47: 863–866. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
