Unraveling amino acid residues critical for allosteric potentiation of (α4)3(β2)2-type nicotinic acetylcholine receptor responses
- PMID: 28446611
- PMCID: PMC5473250
- DOI: 10.1074/jbc.M116.771246
Unraveling amino acid residues critical for allosteric potentiation of (α4)3(β2)2-type nicotinic acetylcholine receptor responses
Abstract
Neuronal nicotinic acetylcholine receptors (nAChRs) are promising drug targets to manage several neurological disorders and nicotine addiction. Growing evidence indicates that positive allosteric modulators of nAChRs improve pharmacological specificity by binding to unique sites present only in a subpopulation of nAChRs. Furthermore, nAChR positive allosteric modulators such as NS9283 and CMPI have been shown to potentiate responses of (α4)3(β2)2 but not (α4)2(β2)3 nAChR isoforms. This selective potentiation underlines that the α4:α4 interface, which is present only in the (α4)3(β2)2 nAChR, is an important and promising drug target. In this report we used site-directed mutagenesis to substitute specific amino acid residues and computational analyses to elucidate CMPI's binding mode at the α4:α4 subunit extracellular interface and identified a unique set of amino acid residues that determined its affinity. We found that amino acid residues α4Gly-41, α4Lys-64, and α4Thr-66 were critical for (α4)3(β2)2 nAChR potentiation by CMPI, but not by NS9283, whereas amino acid substitution at α4His-116, a known determinant of NS9283 and of agonist binding at the α4:α4 subunit interface, did not reduce CMPI potentiation. In contrast, substitutions at α4Gln-124 and α4Thr-126 reduced potentiation by CMPI and NS9283, indicating that their binding sites partially overlap. These results delineate the role of amino acid residues contributing to the α4:α4 subunit extracellular interface in nAChR potentiation. These findings also provide structural information that will facilitate the structure-based design of novel therapeutics that target selectively the (α4)3(β2)2 nAChR.
Keywords: CMPI; NS9283; Positive allosteric modulators; dFBr; drug design; drug development; electrophysiology; ion channel; nicotinic acetylcholine receptors (nAChR); pentameric ligand-gated ion channel.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
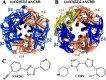







References
-
- Taly A., Corringer P.-J., Guedin D., Lestage P., and Changeux J.-P. (2009) Nicotinic receptors: allosteric transitions and therapeutic targets in the nervous system. Nat. Rev. Drug Discov. 8, 733–750 - PubMed
-
- Hurst R., Rollema H., and Bertrand D. (2013) Nicotinic acetylcholine receptors: from basic science to therapeutics. Pharmacol. Ther. 137, 22–54 - PubMed
-
- Grupe M., Grunnet M., Bastlund J. F., and Jensen A. A. (2015) Targeting α4β2 nicotinic acetylcholine receptors in central nervous system disorders: perspectives on positive allosteric modulation as a therapeutic approach. Basic Clin. Pharmacol. Toxicol. 116, 187–200 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

