Pipeline to Identify Hydroxyproline-Rich Glycoproteins
- PMID: 28446635
- PMCID: PMC5462032
- DOI: 10.1104/pp.17.00294
Pipeline to Identify Hydroxyproline-Rich Glycoproteins
Abstract
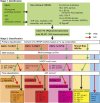
Intrinsically disordered proteins (IDPs) are functional proteins that lack a well-defined three-dimensional structure. The study of IDPs is a rapidly growing area as the crucial biological functions of more of these proteins are uncovered. In plants, IDPs are implicated in plant stress responses, signaling, and regulatory processes. A superfamily of cell wall proteins, the hydroxyproline-rich glycoproteins (HRGPs), have characteristic features of IDPs. Their protein backbones are rich in the disordering amino acid proline, they contain repeated sequence motifs and extensive posttranslational modifications (glycosylation), and they have been implicated in many biological functions. HRGPs are evolutionarily ancient, having been isolated from the protein-rich walls of chlorophyte algae to the cellulose-rich walls of embryophytes. Examination of HRGPs in a range of plant species should provide valuable insights into how they have evolved. Commonly divided into the arabinogalactan proteins, extensins, and proline-rich proteins, in reality, a continuum of structures exists within this diverse and heterogenous superfamily. An inability to accurately classify HRGPs leads to inconsistent gene ontologies limiting the identification of HRGP classes in existing and emerging omics data sets. We present a novel and robust motif and amino acid bias (MAAB) bioinformatics pipeline to classify HRGPs into 23 descriptive subclasses. Validation of MAAB was achieved using available genomic resources and then applied to the 1000 Plants transcriptome project (www.onekp.com) data set. Significant improvement in the detection of HRGPs using multiple-k-mer transcriptome assembly methodology was observed. The MAAB pipeline is readily adaptable and can be modified to optimize the recovery of IDPs from other organisms.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures




References
-
- Bennick A. (1987) Structural and genetic aspects of proline-rich proteins. J Dent Res 66: 457–461 - PubMed
-
- Berthold MR, Cebron N, Dill F, Gabriel TR, Kötter T, Meinl T, Ohl P, Sieb C, Thiel K, Wiswedel B (2008) KNIME: The Konstanz Information Miner. In Preisach C, Burkhardt H, SchmidtThieme L, Decker R, eds, Data Analysis, Machine Learning and Applications. Springer, Berlin, pp 319–326
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

