Insights into the Evolution of Hydroxyproline-Rich Glycoproteins from 1000 Plant Transcriptomes
- PMID: 28446636
- PMCID: PMC5462033
- DOI: 10.1104/pp.17.00295
Insights into the Evolution of Hydroxyproline-Rich Glycoproteins from 1000 Plant Transcriptomes
Abstract
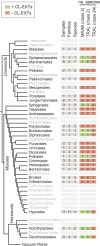
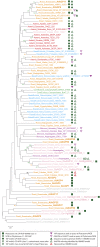
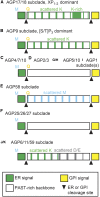
The carbohydrate-rich cell walls of land plants and algae have been the focus of much interest given the value of cell wall-based products to our current and future economies. Hydroxyproline-rich glycoproteins (HRGPs), a major group of wall glycoproteins, play important roles in plant growth and development, yet little is known about how they have evolved in parallel with the polysaccharide components of walls. We investigate the origins and evolution of the HRGP superfamily, which is commonly divided into three major multigene families: the arabinogalactan proteins (AGPs), extensins (EXTs), and proline-rich proteins. Using motif and amino acid bias, a newly developed bioinformatics pipeline, we identified HRGPs in sequences from the 1000 Plants transcriptome project (www.onekp.com). Our analyses provide new insights into the evolution of HRGPs across major evolutionary milestones, including the transition to land and the early radiation of angiosperms. Significantly, data mining reveals the origin of glycosylphosphatidylinositol (GPI)-anchored AGPs in green algae and a 3- to 4-fold increase in GPI-AGPs in liverworts and mosses. The first detection of cross-linking (CL)-EXTs is observed in bryophytes, which suggests that CL-EXTs arose though the juxtaposition of preexisting SPn EXT glycomotifs with refined Y-based motifs. We also detected the loss of CL-EXT in a few lineages, including the grass family (Poaceae), that have a cell wall composition distinct from other monocots and eudicots. A key challenge in HRGP research is tracking individual HRGPs throughout evolution. Using the 1000 Plants output, we were able to find putative orthologs of Arabidopsis pollen-specific GPI-AGPs in basal eudicots.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures




References
-
- Adair WS, Hwang C, Goodenough UW (1983) Identification and visualization of the sexual agglutinin from the mating-type plus flagellar membrane of Chlamydomonas. Cell 33: 183–193 - PubMed
-
- APG IV (2016) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc 181: 1–20
-
- Bacic A, Harris PJ, Stone BA (1988) Structure and function of plant cell walls. In Priess J, ed, The Biochemistry of Plants, Vol 14 Academic Press, New York, pp 297–371
-
- Basile DV, Basile MR (1987) The occurrence of cell wall-associated arabinogalactan proteins in the Hepaticae. Bryologist 90: 401–404
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

