Transcriptomic response of durum wheat to nitrogen starvation
- PMID: 28446759
- PMCID: PMC5430780
- DOI: 10.1038/s41598-017-01377-0
Transcriptomic response of durum wheat to nitrogen starvation
Abstract
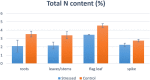
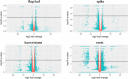
Nitrogen (N) is a key macronutrient representing a limiting factor for plant growth and development and affects productivity in wheat. In this study, durum wheat response to N chronic starvation during grain filling was investigated through a transcriptomic approach in roots, leaves/stems, flag leaf and spikes of cv. Svevo. Nitrogen stress negatively influenced plant height, tillering, flag leaf area, spike and seed traits, and total N content. RNA-seq data revealed 4,626 differentially expressed genes (DEGs). Most transcriptomic changes were observed in roots, with 3,270 DEGs, while 963 were found in leaves/stems, 470 in flag leaf, and 355 in spike tissues. A total of 799 gene ontology (GO) terms were identified, 180 and 619 among the upregulated and downregulated genes, respectively. Among the most addressed GO categories, N compound metabolism, carbon metabolism, and photosynthesis were mostly represented. Interesting DEGs, such as N transporters, genes involved in N assimilation, along with transcription factors, protein kinases and other genes related to stress were highlighted. These results provide valuable information about the transcriptomic response to chronic N stress in durum wheat, which could be useful for future improvement of N use efficiency.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- EPA. Methane and Nitrous Oxide Emissions from Natural Sources. U.S. Environmental Protection Agency. Washington, DC: EPA. 430R10001, https://nepis.epa.gov (2010).
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

