Three-dimensional modeling of single stranded DNA hairpins for aptamer-based biosensors
- PMID: 28446765
- PMCID: PMC5430850
- DOI: 10.1038/s41598-017-01348-5
Three-dimensional modeling of single stranded DNA hairpins for aptamer-based biosensors
Abstract
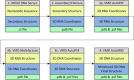
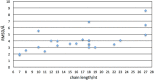
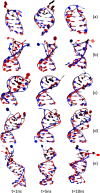
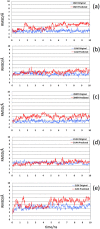
Aptamers consist of short oligonucleotides that bind specific targets. They provide advantages over antibodies, including robustness, low cost, and reusability. Their chemical structure allows the insertion of reporter molecules and surface-binding agents in specific locations, which have been recently exploited for the development of aptamer-based biosensors and direct detection strategies. Mainstream use of these devices, however, still requires significant improvements in optimization for consistency and reproducibility. DNA aptamers are more stable than their RNA counterparts for biomedical applications but have the disadvantage of lacking the wide array of computational tools for RNA structural prediction. Here, we present the first approach to predict from sequence the three-dimensional structures of single stranded (ss) DNA required for aptamer applications, focusing explicitly on ssDNA hairpins. The approach consists of a pipeline that integrates sequentially building ssDNA secondary structure from sequence, constructing equivalent 3D ssRNA models, transforming the 3D ssRNA models into ssDNA 3D structures, and refining the resulting ssDNA 3D structures. Through this pipeline, our approach faithfully predicts the representative structures available in the Nucleic Acid Database and Protein Data Bank databases. Our results, thus, open up a much-needed avenue for integrating DNA in the computational analysis and design of aptamer-based biosensors.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clinical chemistry. 1999;45:1628–1650. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

