MicroRNA and Transcription Factor: Key Players in Plant Regulatory Network
- PMID: 28446918
- PMCID: PMC5388764
- DOI: 10.3389/fpls.2017.00565
MicroRNA and Transcription Factor: Key Players in Plant Regulatory Network
Abstract
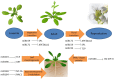
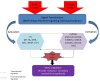
Recent achievements in plant microRNA (miRNA), a large class of small and non-coding RNAs, are very exciting. A wide array of techniques involving forward genetic, molecular cloning, bioinformatic analysis, and the latest technology, deep sequencing have greatly advanced miRNA discovery. A tiny miRNA sequence has the ability to target single/multiple mRNA targets. Most of the miRNA targets are transcription factors (TFs) which have paramount importance in regulating the plant growth and development. Various families of TFs, which have regulated a range of regulatory networks, may assist plants to grow under normal and stress environmental conditions. This present review focuses on the regulatory relationships between miRNAs and different families of TFs like; NF-Y, MYB, AP2, TCP, WRKY, NAC, GRF, and SPL. For instance NF-Y play important role during drought tolerance and flower development, MYB are involved in signal transduction and biosynthesis of secondary metabolites, AP2 regulate the floral development and nodule formation, TCP direct leaf development and growth hormones signaling. WRKY have known roles in multiple stress tolerances, NAC regulate lateral root formation, GRF are involved in root growth, flower, and seed development, and SPL regulate plant transition from juvenile to adult. We also studied the relation between miRNAs and TFs by consolidating the research findings from different plant species which will help plant scientists in understanding the mechanism of action and interaction between these regulators in the plant growth and development under normal and stress environmental conditions.
Keywords: miRNAs; plant development; plant regulators; stress response; transcription factors.
Figures
References
-
- Abe M., Yoshikawa T., Nosaka M., Sakakibara H., Sato Y., Nagato Y., et al. (2010). WAVY LEAF1, an ortholog of Arabidopsis HEN1, regulates shoot development by maintaining MicroRNA and trans-acting small interfering RNA accumulation in rice. Plant Physiol. 154 1335–1346. 10.1104/pp.110.160234 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

