Genome Dynamics of Escherichia coli during Antibiotic Treatment: Transfer, Loss, and Persistence of Genetic Elements In situ of the Infant Gut
- PMID: 28447026
- PMCID: PMC5388698
- DOI: 10.3389/fcimb.2017.00126
Genome Dynamics of Escherichia coli during Antibiotic Treatment: Transfer, Loss, and Persistence of Genetic Elements In situ of the Infant Gut
Abstract
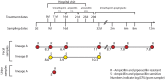
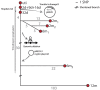
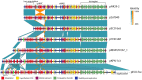
Elucidating the adaptive strategies and plasticity of bacterial genomes in situ is crucial for understanding the epidemiology and evolution of pathogens threatening human health. While much is known about the evolution of Escherichia coli in controlled laboratory environments, less effort has been made to elucidate the genome dynamics of E. coli in its native settings. Here, we follow the genome dynamics of co-existing E. coli lineages in situ of the infant gut during the first year of life. One E. coli lineage causes a urinary tract infection (UTI) and experiences several alterations of its genomic content during subsequent antibiotic treatment. Interestingly, all isolates of this uropathogenic E. coli strain carried a highly stable plasmid implicated in virulence of diverse pathogenic strains from all over the world. While virulence elements are certainly beneficial during infection scenarios, their role in gut colonization and pathogen persistence is poorly understood. We performed in vivo competitive fitness experiments to assess the role of this highly disseminated virulence plasmid in gut colonization, but found no evidence for a direct benefit of plasmid carriage. Through plasmid stability assays, we demonstrate that this plasmid is maintained in a parasitic manner, by strong first-line inheritance mechanisms, acting on the single-cell level, rather than providing a direct survival advantage in the gut. Investigating the ecology of endemic accessory genetic elements, in their pathogenic hosts and native environment, is of vital importance if we want to understand the evolution and persistence of highly virulent and drug resistant bacterial isolates.
Keywords: Escherichia coli; antibiotic treatment; genome evolution; horizontal gene transfer; infant gut; plasmid persistence; urinary tract infections; virulence plasmid dynamics.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

