Transcriptional response of Escherichia coli to ammonia and glucose fluctuations
- PMID: 28447391
- PMCID: PMC5481515
- DOI: 10.1111/1751-7915.12713
Transcriptional response of Escherichia coli to ammonia and glucose fluctuations
Abstract
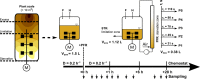
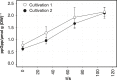
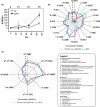
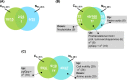
In large-scale production processes, metabolic control is typically achieved by limited supply of essential nutrients such as glucose or ammonia. With increasing bioreactor dimensions, microbial producers such as Escherichia coli are exposed to changing substrate availabilities due to limited mixing. In turn, cells sense and respond to these dynamic conditions leading to frequent activation of their regulatory programmes. Previously, we characterized short- and long-term strategies of cells to adapt to glucose fluctuations. Here, we focused on fluctuating ammonia supply while studying a continuously running two-compartment bioreactor system comprising a stirred tank reactor (STR) and a plug-flow reactor (PFR). The alarmone ppGpp rapidly accumulated in PFR, initiating considerable transcriptional responses after 70 s. About 400 genes were repeatedly switched on/off when E. coli returned to the STR. E. coli revealed highly diverging long-term transcriptional responses in ammonia compared to glucose fluctuations. In contrast, the induction of stringent regulation was a common feature of both short-term responses. Cellular ATP demands for coping with fluctuating ammonia supply were found to increase maintenance by 15%. The identification of genes contributing to the increased ATP demand together with the elucidation of regulatory mechanisms may help to create robust cells and processes for large-scale application.
© 2017 The Authors. Microbial Biotechnology published by John Wiley & Sons Ltd and Society for Applied Microbiology.
Figures

References
-
- Amanullah, A. , McFarlane, C.M. , Emery, A.N. , and Nienow, A.W. (2001) Scale‐down model to simulate spatial pH variations in large‐scale bioreactors. Biotechnol Bioeng 73: 390–399. - PubMed
-
- Benjamini, Y. , and Hochberg, Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57: 289–300.
-
- Buchholz, J. , Graf, M. , Freund, A. , Busche, T. , Kalinowski, J. , Blombach, B. , and Takors, R. (2014) CO2/HCO3 − perturbations of simulated large scale gradients in a scale‐down device cause fast transcriptional responses in Corynebacterium glutamicum . Appl Microbiol Biotechnol 98: 8563–8572. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

