Genome-wide association study of HIV-associated neurocognitive disorder (HAND): A CHARTER group study
- PMID: 28447399
- PMCID: PMC5435520
- DOI: 10.1002/ajmg.b.32530
Genome-wide association study of HIV-associated neurocognitive disorder (HAND): A CHARTER group study
Abstract
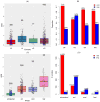
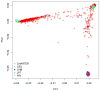
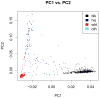
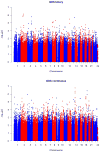
HIV-associated neurocognitive disorder (HAND) often complicates HIV infection despite combination antiretroviral therapy (ART) and may be influenced by host genomics. We performed a genome-wide association study (GWAS) of HAND in 1,050 CNS HIV Anti-Retroviral Therapy Effects Research (CHARTER) Study participants. All participants underwent standardized, comprehensive neurocognitive, and neuromedical assessments to determine if they had cognitive impairment as assessed by the Global Deficit Score (GDS), and individuals with comorbidities that could confound diagnosis of HAND were excluded. Neurocognitive outcomes included GDS-defined neurocognitive impairment (NCI; binary GDS, 366 cases with GDS ≥ 0.5 and 684 controls with GDS < 0.5, and GDS as a continuous variable) and Frascati HAND definitions that incorporate assessment of functional impairment by self-report and performance-based criteria. Genotype data were obtained using the Affymetrix Human SNP Array 6.0 platform. Multivariable logistic or linear regression-based association tests were performed for GDS-defined NCI and HAND. GWAS results did not reveal SNPs meeting the genome-wide significance threshold (5.0 × 10-8 ) for GDS-defined NCI or HAND. For binary GDS, the most significant SNPs were rs6542826 (P = 8.1 × 10-7 ) and rs11681615 (1.2 × 10-6 ), both located on chromosome 2 in SH3RF3. The most significant SNP for continuous GDS was rs11157436 (P = 1.3 × 10-7 ) on chromosome 14 in the T-cell-receptor alpha locus; three other SNPs in this gene were also associated with binary GDS (P ≤ 2.9 × 10-6 ). This GWAS, conducted among ART-era participants from a single cohort with robust neurological phenotyping, suggests roles for several biologically plausible loci in HAND that deserve further exploration. © 2017 Wiley Periodicals, Inc.
Keywords: CHARTER study; GWAS; HIV-associated neurocognitive disorder; genotype; global deficit score; neurocognitive impairment.
© 2017 Wiley Periodicals, Inc.
Figures


References
-
- Antinori A, Arendt G, Becker JT, Brew BJ, Byrd DA, Cherner M, Clifford DB, Cinque P, Epstein LG, Goodkin K, Gisslen M, Grant I, Heaton RK, Joseph J, Marder K, Marra CM, McArthur JC, Nunn M, Price RW, Pulliam L, Robertson KR, Sacktor N, Valcour V, Wojna VE. Updated research nosology for HIV-associated neurocognitive disorders. Neurology. 2007;69(18):1789–1799. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

