PARP3 is a promoter of chromosomal rearrangements and limits G4 DNA
- PMID: 28447610
- PMCID: PMC5414184
- DOI: 10.1038/ncomms15110
PARP3 is a promoter of chromosomal rearrangements and limits G4 DNA
Erratum in
-
Corrigendum: PARP3 is a promoter of chromosomal rearrangements and limits G4 DNA.Nat Commun. 2017 Jun 13;8:15918. doi: 10.1038/ncomms15918. Nat Commun. 2017. PMID: 28607496 Free PMC article.
Abstract
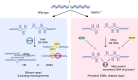
Chromosomal rearrangements are essential events in the pathogenesis of both malignant and nonmalignant disorders, yet the factors affecting their formation are incompletely understood. Here we develop a zinc-finger nuclease translocation reporter and screen for factors that modulate rearrangements in human cells. We identify UBC9 and RAD50 as suppressors and 53BP1, DDB1 and poly(ADP)ribose polymerase 3 (PARP3) as promoters of chromosomal rearrangements across human cell types. We focus on PARP3 as it is dispensable for murine viability and has druggable catalytic activity. We find that PARP3 regulates G quadruplex (G4) DNA in response to DNA damage, which suppresses repair by nonhomologous end-joining and homologous recombination. Chemical stabilization of G4 DNA in PARP3-/- cells leads to widespread DNA double-strand breaks and synthetic lethality. We propose a model in which PARP3 suppresses G4 DNA and facilitates DNA repair by multiple pathways.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





References
-
- Piomboni P., Stendardi A. & Gambera L. Chromosomal aberrations and aneuploidies of spermatozoa. Adv. Exp. Med. Biol. 791, 27–52 (2014). - PubMed
-
- Bochtler T., Frohling S. & Kramer A. Role of chromosomal aberrations in clonal diversity and progression of acute myeloid leukemia. Leukemia 29, 1243–1252 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

