Islet-1 induces the differentiation of mesenchymal stem cells into cardiomyocyte-like cells through the regulation of Gcn5 and DNMT-1
- PMID: 28447752
- PMCID: PMC5428324
- DOI: 10.3892/mmr.2017.6343
Islet-1 induces the differentiation of mesenchymal stem cells into cardiomyocyte-like cells through the regulation of Gcn5 and DNMT-1
Abstract
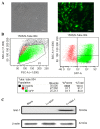
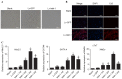
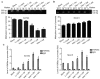
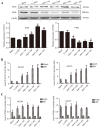
Previous studies from this group demonstrated that insulin gene enhancer binding protein ISL-1 (Islet-1) specifically induces the differentiation of mesenchymal stem cells (MSCs) into cardiomyocyte‑like cells through histone acetylation. However, the underlying mechanisms remain unclear. In the present study, the role of the histone acetylation and DNA methylation on the regulatory mechanism of the Islet‑1 was further investigated by methylation‑specific polymerase chain reaction (PCR), chromatin immunoprecipitation quantitative PCR and western blot analysis. The results demonstrated that Islet‑1 upregulated expression of general control of amino acid biosynthesis protein 5 (Gcn5) and enhanced the binding of Gcn5 to the promoters of GATA binding protein 4 (GATA4) and NK2 homeobox 5 (Nkx2.5). In addition, Islet-1 downregulated DNA methyltransferase (DNMT)‑1 expression and reduced its binding to the GATA4 promoter. In contrast, the amount of DNMT-1 binding on Nkx2.5 did not match the expression trend. Therefore, it was concluded that Islet‑1 may influence the histone acetylation and DNA methylation of GATA4 promoter region via Gcn5 and DNMT‑1 during the MSC differentiation into cardiomyocyte-like cells, thus prompting the expression of GATA4. The Nkx2.5 was likely only affected by histone acetylation instead of DNA methylation. The present study demonstrated that Islet‑1 induces the differentiation of mesenchymal stem cells into cardiomyocyte‑like cells through a specific interaction between histone acetylation and DNA methylation on regulating GATA4.
Figures
Similar articles
-
Islet-1 synergizes with Gcn5 to promote MSC differentiation into cardiomyocytes.Sci Rep. 2020 Feb 4;10(1):1817. doi: 10.1038/s41598-020-58387-8. Sci Rep. 2020. PMID: 32019948 Free PMC article.
-
Islet-1 promotes the cardiac-specific differentiation of mesenchymal stem cells through the regulation of histone acetylation.Int J Mol Med. 2014 May;33(5):1075-82. doi: 10.3892/ijmm.2014.1687. Epub 2014 Mar 6. Int J Mol Med. 2014. PMID: 24604334 Free PMC article.
-
Histone modifications interact with DNA methylation at the GATA4 promoter during differentiation of mesenchymal stem cells into cardiomyocyte-like cells.Cell Prolif. 2016 Jun;49(3):315-29. doi: 10.1111/cpr.12253. Epub 2016 Apr 26. Cell Prolif. 2016. PMID: 27117983 Free PMC article.
-
Islet-1 may function as an assistant factor for histone acetylation and regulation of cardiac development-related transcription factor Mef2c expression.PLoS One. 2013 Oct 17;8(10):e77690. doi: 10.1371/journal.pone.0077690. eCollection 2013. PLoS One. 2013. PMID: 24147056 Free PMC article.
-
Wnt-promoted Isl1 expression through a novel TCF/LEF1 binding site and H3K9 acetylation in early stages of cardiomyocyte differentiation of P19CL6 cells.Mol Cell Biochem. 2014 Jun;391(1-2):183-92. doi: 10.1007/s11010-014-2001-y. Epub 2014 Mar 8. Mol Cell Biochem. 2014. PMID: 24610003
Cited by
-
Smyd1 Orchestrates Early Heart Development Through Positive and Negative Gene Regulation.Front Cell Dev Biol. 2021 Apr 1;9:654682. doi: 10.3389/fcell.2021.654682. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33869215 Free PMC article.
-
Optimizing Cardiomyocyte Differentiation: Comparative Analysis of Bone Marrow and Adipose-Derived Mesenchymal Stem Cells in Rats Using 5-Azacytidine and Low-Dose FGF and IGF Treatment.Biomedicines. 2024 Aug 22;12(8):1923. doi: 10.3390/biomedicines12081923. Biomedicines. 2024. PMID: 39200387 Free PMC article.
-
Human cardiomyocyte-derived exosomes induce cardiac gene expressions in mesenchymal stromal cells within 3D hyaluronic acid hydrogels and in dose-dependent manner.J Mater Sci Mater Med. 2021 Jan 19;32(1):2. doi: 10.1007/s10856-020-06474-7. J Mater Sci Mater Med. 2021. PMID: 33469781 Free PMC article.
-
Exploring the Potential Effects of Cryopreservation on the Biological Characteristics and Cardiomyogenic Differentiation of Rat Adipose-Derived Mesenchymal Stem Cells.Int J Mol Sci. 2024 Sep 13;25(18):9908. doi: 10.3390/ijms25189908. Int J Mol Sci. 2024. PMID: 39337396 Free PMC article.
-
Histone Acetyltransferases and Stem Cell Identity.Cancers (Basel). 2021 May 17;13(10):2407. doi: 10.3390/cancers13102407. Cancers (Basel). 2021. PMID: 34067525 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

