Host-selected mutations converging on a global regulator drive an adaptive leap towards symbiosis in bacteria
- PMID: 28447935
- PMCID: PMC5466423
- DOI: 10.7554/eLife.24414
Host-selected mutations converging on a global regulator drive an adaptive leap towards symbiosis in bacteria
Abstract
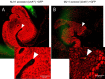
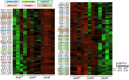
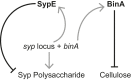
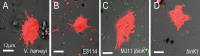
Host immune and physical barriers protect against pathogens but also impede the establishment of essential symbiotic partnerships. To reveal mechanisms by which beneficial organisms adapt to circumvent host defenses, we experimentally evolved ecologically distinct bioluminescent Vibrio fischeri by colonization and growth within the light organs of the squid Euprymna scolopes. Serial squid passaging of bacteria produced eight distinct mutations in the binK sensor kinase gene, which conferred an exceptional selective advantage that could be demonstrated through both empirical and theoretical analysis. Squid-adaptive binK alleles promoted colonization and immune evasion that were mediated by cell-associated matrices including symbiotic polysaccharide (Syp) and cellulose. binK variation also altered quorum sensing, raising the threshold for luminescence induction. Preexisting coordinated regulation of symbiosis traits by BinK presented an efficient solution where altered BinK function was the key to unlock multiple colonization barriers. These results identify a genetic basis for microbial adaptability and underscore the importance of hosts as selective agents that shape emergent symbiont populations.
Keywords: Euprymna scolopes; Vibrio fischeri; evolutionary biology; experimental evolution; genomics; infectious disease; microbiology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures












References
-
- Altura MA, Heath-Heckman EA, Gillette A, Kremer N, Krachler AM, Brennan C, Ruby EG, Orth K, McFall-Ngai MJ. The first engagement of partners in the Euprymna scolopes-Vibrio fischeri symbiosis is a two-step process initiated by a few environmental symbiont cells. Environmental Microbiology. 2013;15:2937–2950. doi: 10.1111/1462-2920.12179. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

