Role of Archaeal HerA Protein in the Biology of the Bacterium Thermus thermophilus
- PMID: 28448436
- PMCID: PMC5448004
- DOI: 10.3390/genes8050130
Role of Archaeal HerA Protein in the Biology of the Bacterium Thermus thermophilus
Abstract
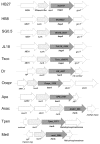
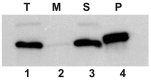
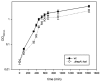
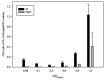
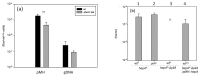
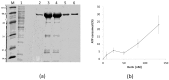
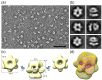
Intense gene flux between prokaryotes result in high percentage of archaeal genes in the genome of the thermophilic bacteria Thermus spp. Among these archaeal genes a homolog to the Sulfolobus spp. HerA protein appears in all of the Thermus spp. strains so far sequenced (HepA). The role of HepA in Thermus thermophilus HB27 has been analyzed using deletion mutants, and its structure resolved at low resolution by electron microscopy. Recombinant HepA shows DNA-dependent ATPase activity and its structure revealed a double ring, conically-shaped hexamer with an upper diameter of 150 Å and a bottom module of 95 Å. A central pore was detected in the structure that ranges from 13 Å at one extreme, to 30 Å at the other. Mutants lacking HepA show defective natural competence and DNA donation capability in a conjugation-like process termed "transjugation", and also high sensitivity to UV and dramatic sensitivity to high temperatures. These data support that acquisition of an ancestral archaeal HerA has been fundamental for the adaptation of Thermus spp. to high temperatures.
Keywords: DNA repair; HerA; Thermus; hexameric ATPase; lateral gene transfer; transformation; transjugation.
Conflict of interest statement
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures

References
-
- Omelchenko M.V., Wolf Y.I., Gaidamakova E.K., Matrosova V.Y., Vasilenko A., Zhai M., Daly M.J., Koonin E.V., Makarova K.S. Comparative genomics of thermus thermophilus and deinococcus radiodurans: Divergent routes of adaptation to thermophily and radiation resistance. BMC Evol. Biol. 2005;5:57. doi: 10.1186/1471-2148-5-57. - DOI - PMC - PubMed
-
- Iyer L.M., Makarova K.S., Koonin E.V., Aravind L. Comparative genomics of the ftsk—HerA superfamily of pumping atpases: Implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32:5260–5279. doi: 10.1093/nar/gkh828. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

