Suppression of the Escherichia coli dnaA46 mutation by changes in the activities of the pyruvate-acetate node links DNA replication regulation to central carbon metabolism
- PMID: 28448512
- PMCID: PMC5407757
- DOI: 10.1371/journal.pone.0176050
Suppression of the Escherichia coli dnaA46 mutation by changes in the activities of the pyruvate-acetate node links DNA replication regulation to central carbon metabolism
Abstract
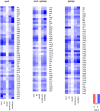
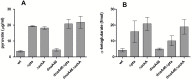
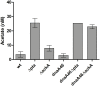
To ensure faithful transmission of genetic material to progeny cells, DNA replication is tightly regulated, mainly at the initiation step. Escherichia coli cells regulate the frequency of initiation according to growth conditions. Results of the classical, as well as the latest studies, suggest that the DNA replication in E. coli starts at a predefined, constant cell volume per chromosome but the mechanisms coordinating DNA replication with cell growth are still not fully understood. Results of recent investigations have revealed a role of metabolic pathway proteins in the control of cell division and a direct link between metabolism and DNA replication has also been suggested both in Bacillus subtilis and E. coli cells. In this work we show that defects in the acetate overflow pathway suppress the temperature-sensitivity of a defective replication initiator-DnaA under acetogenic growth conditions. Transcriptomic and metabolic analyses imply that this suppression is correlated with pyruvate accumulation, resulting from alterations in the pyruvate dehydrogenase (PDH) activity. Consequently, deletion of genes encoding the pyruvate dehydrogenase subunits likewise resulted in suppression of the thermal-sensitive growth of the dnaA46 strain. We propose that the suppressor effect may be directly related to the PDH complex activity, providing a link between an enzyme of the central carbon metabolism and DNA replication.
Conflict of interest statement
Figures






Similar articles
-
The nucleoid protein H-NS facilitates chromosome DNA replication in Escherichia coli dnaA mutants.J Bacteriol. 1996 Oct;178(19):5790-2. doi: 10.1128/jb.178.19.5790-5792.1996. J Bacteriol. 1996. PMID: 8824628 Free PMC article.
-
Physiological response of central metabolism in Escherichia coli to deletion of pyruvate oxidase and introduction of heterologous pyruvate carboxylase.Biotechnol Bioeng. 2005 Apr 5;90(1):64-76. doi: 10.1002/bit.20418. Biotechnol Bioeng. 2005. PMID: 15736164
-
Central carbon metabolism influences fidelity of DNA replication in Escherichia coli.Mutat Res. 2012 Mar 1;731(1-2):99-106. doi: 10.1016/j.mrfmmm.2011.12.005. Epub 2011 Dec 17. Mutat Res. 2012. PMID: 22198407
-
Regulation of initiation of Bacillus subtilis chromosome replication.Plasmid. 1999 Jan;41(1):17-29. doi: 10.1006/plas.1998.1381. Plasmid. 1999. PMID: 9887303 Review.
-
Replicating DNA by cell factories: roles of central carbon metabolism and transcription in the control of DNA replication in microbes, and implications for understanding this process in human cells.Microb Cell Fact. 2013 May 29;12:55. doi: 10.1186/1475-2859-12-55. Microb Cell Fact. 2013. PMID: 23714207 Free PMC article. Review.
Cited by
-
Multiple links connect central carbon metabolism to DNA replication initiation and elongation in Bacillus subtilis.DNA Res. 2018 Dec 1;25(6):641-653. doi: 10.1093/dnares/dsy031. DNA Res. 2018. PMID: 30256918 Free PMC article.
-
The metabolic control of DNA replication: mechanism and function.Open Biol. 2023 Aug;13(8):230220. doi: 10.1098/rsob.230220. Epub 2023 Aug 16. Open Biol. 2023. PMID: 37582405 Free PMC article. Review.
-
Pyruvate kinase, a metabolic sensor powering glycolysis, drives the metabolic control of DNA replication.BMC Biol. 2022 Apr 13;20(1):87. doi: 10.1186/s12915-022-01278-3. BMC Biol. 2022. PMID: 35418203 Free PMC article.
-
Open Questions about the Roles of DnaA, Related Proteins, and Hyperstructure Dynamics in the Cell Cycle.Life (Basel). 2023 Sep 10;13(9):1890. doi: 10.3390/life13091890. Life (Basel). 2023. PMID: 37763294 Free PMC article. Review.
-
Bacterial cell proliferation: from molecules to cells.FEMS Microbiol Rev. 2021 Jan 8;45(1):fuaa046. doi: 10.1093/femsre/fuaa046. FEMS Microbiol Rev. 2021. PMID: 32990752 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

