Localization of adenovirus morphogenesis players, together with visualization of assembly intermediates and failed products, favor a model where assembly and packaging occur concurrently at the periphery of the replication center
- PMID: 28448571
- PMCID: PMC5409498
- DOI: 10.1371/journal.ppat.1006320
Localization of adenovirus morphogenesis players, together with visualization of assembly intermediates and failed products, favor a model where assembly and packaging occur concurrently at the periphery of the replication center
Abstract
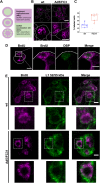
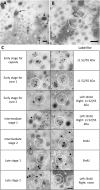
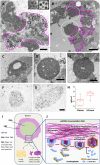
Adenovirus (AdV) morphogenesis is a complex process, many aspects of which remain unclear. In particular, it is not settled where in the nucleus assembly and packaging occur, and whether these processes occur in a sequential or a concerted manner. Here we use immunofluorescence and immunoelectron microscopy (immunoEM) to trace packaging factors and structural proteins at late times post infection by either wildtype virus or a delayed packaging mutant. We show that representatives of all assembly factors are present in the previously recognized peripheral replicative zone, which therefore is the AdV assembly factory. Assembly intermediates and abortive products observed in this region favor a concurrent assembly and packaging model comprising two pathways, one for capsid proteins and another one for core components. Only when both pathways are coupled by correct interaction between packaging proteins and the genome is the viral particle produced. Decoupling generates accumulation of empty capsids and unpackaged cores.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Adenovirus Core Proteins: Structure and Function.Viruses. 2021 Feb 28;13(3):388. doi: 10.3390/v13030388. Viruses. 2021. PMID: 33671079 Free PMC article. Review.
-
The adenovirus major core protein VII is dispensable for virion assembly but is essential for lytic infection.PLoS Pathog. 2017 Jun 19;13(6):e1006455. doi: 10.1371/journal.ppat.1006455. eCollection 2017 Jun. PLoS Pathog. 2017. PMID: 28628648 Free PMC article.
-
Mysteries of adenovirus packaging.J Virol. 2025 May 20;99(5):e0018025. doi: 10.1128/jvi.00180-25. Epub 2025 Apr 17. J Virol. 2025. PMID: 40243339 Free PMC article. Review.
-
Formation of adenovirus DNA replication compartments and viral DNA accumulation sites by host chromatin regulatory proteins including NPM1.FEBS J. 2020 Jan;287(1):205-217. doi: 10.1111/febs.15027. Epub 2019 Aug 9. FEBS J. 2020. PMID: 31365788
-
Altering the Ad5 packaging domain affects the maturation of the Ad particles.PLoS One. 2011;6(5):e19564. doi: 10.1371/journal.pone.0019564. Epub 2011 May 18. PLoS One. 2011. PMID: 21611162 Free PMC article.
Cited by
-
Human Adenovirus Gene Expression and Replication Is Regulated through Dynamic Changes in Nucleoprotein Structure throughout Infection.Viruses. 2023 Jan 5;15(1):161. doi: 10.3390/v15010161. Viruses. 2023. PMID: 36680201 Free PMC article. Review.
-
RANBP2 and USP9x regulate nuclear import of adenovirus minor coat protein IIIa.PLoS Pathog. 2022 Jun 16;18(6):e1010588. doi: 10.1371/journal.ppat.1010588. eCollection 2022 Jun. PLoS Pathog. 2022. PMID: 35709296 Free PMC article.
-
The human adenovirus type 5 E1B 55kDa protein interacts with RNA promoting timely DNA replication and viral late mRNA metabolism.PLoS One. 2019 Apr 3;14(4):e0214882. doi: 10.1371/journal.pone.0214882. eCollection 2019. PLoS One. 2019. PMID: 30943256 Free PMC article.
-
In vitro methods for testing antiviral drugs.Biotechnol Adv. 2018 May-Jun;36(3):557-576. doi: 10.1016/j.biotechadv.2017.12.016. Epub 2017 Dec 29. Biotechnol Adv. 2018. PMID: 29292156 Free PMC article. Review.
-
Adenovirus Core Proteins: Structure and Function.Viruses. 2021 Feb 28;13(3):388. doi: 10.3390/v13030388. Viruses. 2021. PMID: 33671079 Free PMC article. Review.
References
-
- Pérez-Berná AJ, Marion S, Chichón FJ, Fernández JJ, Winkler DC, Carrascosa JL, et al. Distribution of DNA-condensing protein complexes in the adenovirus core. Nucleic Acids Res. 2015;43(8):4274–83. doi: 10.1093/nar/gkv187 - DOI - PMC - PubMed
-
- San Martín C. Latest Insights on Adenovirus Structure and Assembly. Viruses. 2012;4(5):847–77. doi: 10.3390/v4050847 - DOI - PMC - PubMed
-
- Liu H, Jin L, Koh SB, Atanasov I, Schein S, Wu L, et al. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science. 2010;329(5995):1038–43. Epub 2010/08/28. doi: 10.1126/science.1187433 - DOI - PMC - PubMed
-
- Ishibashi M, Maizel JV Jr. The polypeptides of adenovirus. V. Young virions, structural intermediate between top components and aged virions. Virology. 1974;57(2):409–24. Epub 1974/02/01. - PubMed
-
- Mangel WF, San Martín C. Structure, function and dynamics in adenovirus maturation. Viruses. 2014;6(11):4536–70. PubMed Central PMCID: PMC4246237. doi: 10.3390/v6114536 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

