Circular RNAs with hammerhead ribozymes encoded in eukaryotic genomes: The enemy at home
- PMID: 28448743
- PMCID: PMC5680766
- DOI: 10.1080/15476286.2017.1321730
Circular RNAs with hammerhead ribozymes encoded in eukaryotic genomes: The enemy at home
Abstract
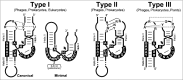
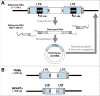
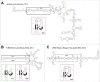
A new family of non-autonomous retrotransposons with self-cleaving hammerhead ribozymes, the so called retrozymes, has recently been found encoded in diverse plant genomes. These retroelements can be actively transcribed, and their RNAs accumulate in the cells as abundant non-coding circular RNAs (circRNAs) of small size (600-1000 nt). Related circRNAs with self-cleaving ribozymes had already been described in plants, and belong to a group of infectious RNA agents with an uncertain origin: the viroids and viroid-like satellites of plant RNA viruses. These pathogenic circRNAs show many structural similarities with retrozyme circRNAs, and both have been found to occur in flowering plants as heterogeneous RNA molecules of positive and negative polarities. Taking all these data together, we hypothesize that circRNAs encoded by genomic retrozymes could have given origin to infectious circRNAs with self-cleaving ribozymes. Moreover, we propose that retrozymes in time could have evolved from the ancient family of Penelope-like retroelements, which also harbour hammerhead ribozymes. Putative retrozyme sequences with hammerhead ribozymes have been detected as well in metazoan genomes, opening the door to a common occurrence of circRNAs with self-cleaving motifs among eukaryotes.
Keywords: Circular RNA; retrotransposons; ribozyme; viral satellite RNA; viroid.
Figures
References
-
- Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 1982; 31:147-57; PMID:6297745; https://doi.org/ 10.1016/0092-8674(82)90414-7 - DOI - PubMed
-
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 1983; 35:849-57; PMID:6197186; https://doi.org/ 10.1016/0092-8674(83)90117-4 - DOI - PubMed
-
- Crick FH. The origin of the genetic code. J Mol Biol 1968; 38:367-79; PMID:4887876; https://doi.org/ 10.1016/0022-2836(68)90392-6 - DOI - PubMed
-
- Orgel LE. Evolution of the genetic apparatus. J Mol Biol 1968; 38:381-93; PMID:5718557; https://doi.org/ 10.1016/0022-2836(68)90393-8 - DOI - PubMed
-
- Woese CR. The fundamental nature of the genetic code: Prebiotic interactions between polynucleotides and polyamino acids or their derivatives. Proc Natl Acad Sci U S A 1968; 59:110-7; PMID:5242115; https://doi.org/ 10.1073/pnas.59.1.110 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
