Calculating the quality of public high-throughput sequencing data to obtain a suitable subset for reanalysis from the Sequence Read Archive
- PMID: 28449062
- PMCID: PMC5459929
- DOI: 10.1093/gigascience/gix029
Calculating the quality of public high-throughput sequencing data to obtain a suitable subset for reanalysis from the Sequence Read Archive
Abstract
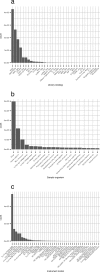
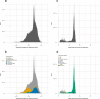
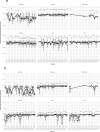
It is important for public data repositories to promote the reuse of archived data. In the growing field of omics science, however, the increasing number of submissions of high-throughput sequencing (HTSeq) data to public repositories prevents users from choosing a suitable data set from among the large number of search results. Repository users need to be able to set a threshold to reduce the number of results to obtain a suitable subset of high-quality data for reanalysis. We calculated the quality of sequencing data archived in a public data repository, the Sequence Read Archive (SRA), by using the quality control software FastQC. We obtained quality values for 1 171 313 experiments, which can be used to evaluate the suitability of data for reuse. We also visualized the data distribution in SRA by integrating the quality information and metadata of experiments and samples. We provide quality information of all of the archived sequencing data, which enable users to obtain sufficient quality sequencing data for reanalyses. The calculated quality data are available to the public in various formats. Our data also provide an example of enhancing the reuse of public data by adding metadata to published research data by a third party.
Keywords: high-throughput sequencing; sequencing quality; public data; database.
© The Authors 2017. Published by Oxford University Press.
Figures

References
-
- Organisation for Economic Co-operation and Development OECD Principles and Guidelines for Access to Research Data from Public Funding. Paris: OECD; 2007. http://www.oecd.org/science/sci-tech/38500813.pdf. (11 November 2016, date last accessed).
-
- Ball CA, Sherlock G, Brazma A. Funding high-throughput data sharing. Nat Biotechnol 2004;22(9):1179–83. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

