Two ribosome recruitment sites direct multiple translation events within HIV1 Gag open reading frame
- PMID: 28449096
- PMCID: PMC5499600
- DOI: 10.1093/nar/gkx303
Two ribosome recruitment sites direct multiple translation events within HIV1 Gag open reading frame
Erratum in
-
Two ribosome recruitment sites direct multiple translation events within HIV1 Gag open reading frame.Nucleic Acids Res. 2017 Jul 7;45(12):7538. doi: 10.1093/nar/gkx401. Nucleic Acids Res. 2017. PMID: 28472379 Free PMC article. No abstract available.
Abstract
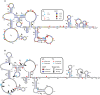
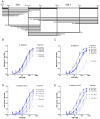
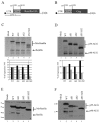
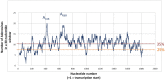
In the late phase of the HIV virus cycle, the unspliced genomic RNA is exported to the cytoplasm for the necessary translation of the Gag and Gag-pol polyproteins. Three distinct translation initiation mechanisms ensuring Gag production have been described with little rationale for their multiplicity. The Gag-IRES has the singularity to be located within Gag ORF and to directly interact with ribosomal 40S. Aiming at elucidating the specificity and the relevance of this interaction, we probed HIV-1 Gag-IRES structure and developed an innovative integrative modelling strategy to take into account all the gathered information. We propose a novel Gag-IRES secondary structure strongly supported by all experimental data. We further demonstrate the presence of two regions within Gag-IRES that independently and directly interact with the ribosome. Importantly, these binding sites are functionally relevant to Gag translation both in vitro and ex vivo. This work provides insight into the Gag-IRES molecular mechanism and gives compelling evidence for its physiological importance. It allows us to propose original hypotheses about the IRES physiological role and conservation among primate lentiviruses.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
A conserved structure within the HIV gag open reading frame that controls translation initiation directly recruits the 40S subunit and eIF3.Nucleic Acids Res. 2011 Mar;39(6):2367-77. doi: 10.1093/nar/gkq1118. Epub 2010 Nov 11. Nucleic Acids Res. 2011. PMID: 21071421 Free PMC article.
-
A new type of IRES within gag coding region recruits three initiation complexes on HIV-2 genomic RNA.Nucleic Acids Res. 2010 Mar;38(4):1367-81. doi: 10.1093/nar/gkp1109. Epub 2009 Dec 6. Nucleic Acids Res. 2010. PMID: 19969542 Free PMC article.
-
Cell type specificity and structural determinants of IRES activity from the 5' leaders of different HIV-1 transcripts.Nucleic Acids Res. 2013 Jul;41(13):6698-714. doi: 10.1093/nar/gkt358. Epub 2013 May 9. Nucleic Acids Res. 2013. PMID: 23661682 Free PMC article.
-
Deconstructing internal ribosome entry site elements: an update of structural motifs and functional divergences.Open Biol. 2018 Nov 28;8(11):180155. doi: 10.1098/rsob.180155. Open Biol. 2018. PMID: 30487301 Free PMC article. Review.
-
tRNA-mimicry in IRES-mediated translation and recoding.RNA Biol. 2016 Nov;13(11):1068-1074. doi: 10.1080/15476286.2016.1219833. Epub 2016 Aug 11. RNA Biol. 2016. PMID: 27654067 Free PMC article. Review.
Cited by
-
sRNA-controlled iron sparing response in Staphylococci.Nucleic Acids Res. 2022 Aug 26;50(15):8529-8546. doi: 10.1093/nar/gkac648. Nucleic Acids Res. 2022. PMID: 35904807 Free PMC article.
-
HIV-1 tolerates changes in A-count in a small segment of the pol gene.Retrovirology. 2017 Sep 5;14(1):43. doi: 10.1186/s12977-017-0367-0. Retrovirology. 2017. PMID: 28870251 Free PMC article.
-
SHAPE Probing to Screen Computationally Designed RNA.Methods Mol Biol. 2025;2847:177-191. doi: 10.1007/978-1-0716-4079-1_12. Methods Mol Biol. 2025. PMID: 39312144
-
IPANEMAP Suite: a pipeline for probing-informed RNA structure modeling.NAR Genom Bioinform. 2025 Mar 25;7(1):lqaf028. doi: 10.1093/nargab/lqaf028. eCollection 2025 Mar. NAR Genom Bioinform. 2025. PMID: 40134455 Free PMC article.
-
Insights into Structural and Mechanistic Features of Viral IRES Elements.Front Microbiol. 2018 Jan 4;8:2629. doi: 10.3389/fmicb.2017.02629. eCollection 2017. Front Microbiol. 2018. PMID: 29354113 Free PMC article. Review.
References
-
- Pelletier J., Sonenberg N.. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988; 334:320–325. - PubMed
-
- Lomakin I.B., Hellen C.U., Pestova T.V.. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 2000; 20:6019–6029. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

