Integrated analysis of individual codon contribution to protein biosynthesis reveals a new approach to improving the basis of rational gene design
- PMID: 28449100
- PMCID: PMC5737324
- DOI: 10.1093/dnares/dsx014
Integrated analysis of individual codon contribution to protein biosynthesis reveals a new approach to improving the basis of rational gene design
Abstract
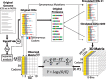
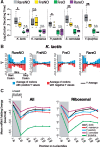
Gene codon optimization may be impaired by the misinterpretation of frequency and optimality of codons. Although recent studies have revealed the effects of codon usage bias (CUB) on protein biosynthesis, an integrated perspective of the biological role of individual codons remains unknown. Unlike other previous studies, we show, through an integrated framework that attributes of codons such as frequency, optimality and positional dependency should be combined to unveil individual codon contribution for protein biosynthesis. We designed a codon quantification method for assessing CUB as a function of position within genes with a novel constraint: the relativity of position-dependent codon usage shaped by coding sequence length. Thus, we propose a new way of identifying the enrichment, depletion and non-uniform positional distribution of codons in different regions of yeast genes. We clustered codons that shared attributes of frequency and optimality. The cluster of non-optimal codons with rare occurrence displayed two remarkable characteristics: higher codon decoding time than frequent-non-optimal cluster and enrichment at the 5'-end region, where optimal codons with the highest frequency are depleted. Interestingly, frequent codons with non-optimal adaptation to tRNAs are uniformly distributed in the Saccharomyces cerevisiae genes, suggesting their determinant role as a speed regulator in protein elongation.
Keywords: codon usage bias; microbial biotechnology; position-dependent codon usage; rational gene design; yeast genomics.
© The Author 2017. Published by Oxford University Press on behalf of Kazusa DNA Research Institute.
Figures






Similar articles
-
Coevolution of codon usage and transfer RNA abundance.Nature. 1987 Feb 19-25;325(6106):728-30. doi: 10.1038/325728a0. Nature. 1987. PMID: 2434856
-
Adjacent Codons Act in Concert to Modulate Translation Efficiency in Yeast.Cell. 2016 Jul 28;166(3):679-690. doi: 10.1016/j.cell.2016.05.070. Epub 2016 Jun 30. Cell. 2016. PMID: 27374328 Free PMC article.
-
Codon optimality is a major determinant of mRNA stability.Cell. 2015 Mar 12;160(6):1111-24. doi: 10.1016/j.cell.2015.02.029. Cell. 2015. PMID: 25768907 Free PMC article.
-
The Yin and Yang of codon usage.Hum Mol Genet. 2016 Oct 1;25(R2):R77-R85. doi: 10.1093/hmg/ddw207. Epub 2016 Jun 27. Hum Mol Genet. 2016. PMID: 27354349 Free PMC article. Review.
-
Codon usage and tRNA content in unicellular and multicellular organisms.Mol Biol Evol. 1985 Jan;2(1):13-34. doi: 10.1093/oxfordjournals.molbev.a040335. Mol Biol Evol. 1985. PMID: 3916708 Review.
Cited by
-
Interplay between Position-Dependent Codon Usage Bias and Hydrogen Bonding at the 5' End of ORFeomes.mSystems. 2020 Aug 11;5(4):e00613-20. doi: 10.1128/mSystems.00613-20. mSystems. 2020. PMID: 32788408 Free PMC article.
-
Mechanisms governing codon usage bias and the implications for protein expression in the chloroplast of Chlamydomonas reinhardtii.Plant J. 2022 Nov;112(4):919-945. doi: 10.1111/tpj.15970. Epub 2022 Oct 19. Plant J. 2022. PMID: 36071273 Free PMC article. Review.
-
Genomic Evidence for Simultaneous Optimization of Transcription and Translation through Codon Variants in the pmoCAB Operon of Type Ia Methanotrophs.mSystems. 2019 Jul 23;4(4):e00342-19. doi: 10.1128/mSystems.00342-19. mSystems. 2019. PMID: 31337658 Free PMC article.
-
Standard Intein Gene Expression Ramps (SIGER) for Protein-Independent Expression Control.ACS Synth Biol. 2023 Apr 21;12(4):1058-1071. doi: 10.1021/acssynbio.2c00530. Epub 2023 Mar 15. ACS Synth Biol. 2023. PMID: 36920366 Free PMC article.
References
-
- Ikemura T. 1981, Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes, J. Mol. Biol., 146, 1–21. - PubMed
-
- Dong H., Nilsson L., Kurland C.G.. 1996, Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates, J. Mol. Biol., 260, 649–63. - PubMed
-
- Hahn M.W., Mezey J.G., Begun D.J., et al.2005, Evolutionary genomics: codon bias and selection on single genomes, Nature, 433, E5–6. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

