Autophagy compensates impaired energy metabolism in CLPXP-deficient Podospora anserina strains and extends healthspan
- PMID: 28449241
- PMCID: PMC5506401
- DOI: 10.1111/acel.12600
Autophagy compensates impaired energy metabolism in CLPXP-deficient Podospora anserina strains and extends healthspan
Abstract
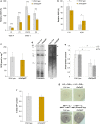
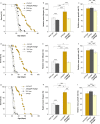
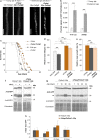
The degradation of nonfunctional mitochondrial proteins is of fundamental relevance for maintenance of cellular homeostasis. The heteromeric CLPXP protein complex in the mitochondrial matrix is part of this process. In the fungal aging model Podospora anserina, ablation of CLPXP leads to an increase in healthy lifespan. Here, we report that this counterintuitive increase depends on a functional autophagy machinery. In PaClpXP mutants, autophagy is involved in energy conservation and the compensation of impairments in respiration. Strikingly, despite the impact on mitochondrial function, it is not mitophagy but general autophagy that is constitutively induced and required for longevity. In contrast, in another long-lived mutant ablated for the mitochondrial PaIAP protease, autophagy is neither induced nor required for lifespan extension. Our data provide novel mechanistic insights into the capacity of different forms of autophagy to compensate impairments of specific components of the complex mitochondrial quality control network and about the biological role of mitochondrial CLPXP in the control of cellular energy metabolism.
Keywords: Podospora anserina; CLPXP protease; aging; autophagy; energy metabolism; mitochondria.
© 2017 The Authors. Aging Cell published by the Anatomical Society and John Wiley & Sons Ltd.
Figures


Similar articles
-
Identification of potential mitochondrial CLPXP protease interactors and substrates suggests its central role in energy metabolism.Sci Rep. 2015 Dec 17;5:18375. doi: 10.1038/srep18375. Sci Rep. 2015. PMID: 26679294 Free PMC article.
-
Human CLPP reverts the longevity phenotype of a fungal ClpP deletion strain.Nat Commun. 2013;4:1397. doi: 10.1038/ncomms2397. Nat Commun. 2013. PMID: 23360988 Free PMC article.
-
Proteomic analysis of mitochondria from senescent Podospora anserina casts new light on ROS dependent aging mechanisms.Exp Gerontol. 2014 Aug;56:13-25. doi: 10.1016/j.exger.2014.02.008. Epub 2014 Feb 18. Exp Gerontol. 2014. PMID: 24556281
-
Cell death by incompatibility in the fungus Podospora.Semin Cancer Biol. 2007 Apr;17(2):101-11. doi: 10.1016/j.semcancer.2006.11.009. Epub 2006 Dec 15. Semin Cancer Biol. 2007. PMID: 17204431 Review.
-
Mitochondrial metabolism and aging in the filamentous fungus Podospora anserina.Biochim Biophys Acta. 2006 May-Jun;1757(5-6):604-10. doi: 10.1016/j.bbabio.2006.03.005. Epub 2006 Mar 30. Biochim Biophys Acta. 2006. PMID: 16624249 Review.
Cited by
-
A Network of Pathways Controlling Cellular Homeostasis Affects the Onset of Senescence in Podospora anserina.J Fungi (Basel). 2021 Mar 31;7(4):263. doi: 10.3390/jof7040263. J Fungi (Basel). 2021. PMID: 33807190 Free PMC article. Review.
-
Characterization of TR-107, a novel chemical activator of the human mitochondrial protease ClpP.Pharmacol Res Perspect. 2022 Aug;10(4):e00993. doi: 10.1002/prp2.993. Pharmacol Res Perspect. 2022. PMID: 35929764 Free PMC article.
-
Guidelines and recommendations on yeast cell death nomenclature.Microb Cell. 2018 Jan 1;5(1):4-31. doi: 10.15698/mic2018.01.607. Microb Cell. 2018. PMID: 29354647 Free PMC article. Review.
-
Mitochondrial Phospholipid Homeostasis Is Regulated by the i-AAA Protease PaIAP and Affects Organismic Aging.Cells. 2021 Oct 16;10(10):2775. doi: 10.3390/cells10102775. Cells. 2021. PMID: 34685755 Free PMC article.
-
Identification and immune features of cuproptosis-related molecular clusters in polycystic ovary syndrome.Sci Rep. 2023 Jan 18;13(1):980. doi: 10.1038/s41598-022-27326-0. Sci Rep. 2023. PMID: 36653385 Free PMC article.
References
-
- Al‐Furoukh N, Ianni A, Nolte H, Holper S, Kruger M, Wanrooij S, Braun T (2015) ClpX stimulates the mitochondrial unfolded protein response (UPRmt) in mammalian cells. Biochim. Biophys. Acta 1853, 2580–2591. - PubMed
-
- Brust D, Daum B, Breunig C, Hamann A, Kühlbrandt W, Osiewacz HD (2010) Cyclophilin D links programmed cell death and organismal aging in Podospora anserina . Aging Cell 9, 761–775. - PubMed
-
- Cole A, Wang Z, Coyaud E, Voisin V, Gronda M, Jitkova Y, Mattson R, Hurren R, Babovic S, Maclean N, Restall I, Wang X, Jeyaraju DV, Sukhai MA, Prabha S, Bashir S, Ramakrishnan A, Leung E, Qia YH, Zhang N, Combes KR, Ketela T, Lin F, Houry WA, Aman A, Al‐Awar R, Zheng W, Wienholds E, Xu CJ, Dick J, Wang JC, Moffat J, Minden MD, Eaves CJ, Bader GD, Hao Z, Kornblau SM, Raught B, Schimmer AD (2015) Inhibition of the mitochondrial protease ClpP as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell 27, 864–876. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

