CloVR-Comparative: automated, cloud-enabled comparative microbial genome sequence analysis pipeline
- PMID: 28449639
- PMCID: PMC5408420
- DOI: 10.1186/s12864-017-3717-3
CloVR-Comparative: automated, cloud-enabled comparative microbial genome sequence analysis pipeline
Abstract
Background: The benefit of increasing genomic sequence data to the scientific community depends on easy-to-use, scalable bioinformatics support. CloVR-Comparative combines commonly used bioinformatics tools into an intuitive, automated, and cloud-enabled analysis pipeline for comparative microbial genomics.
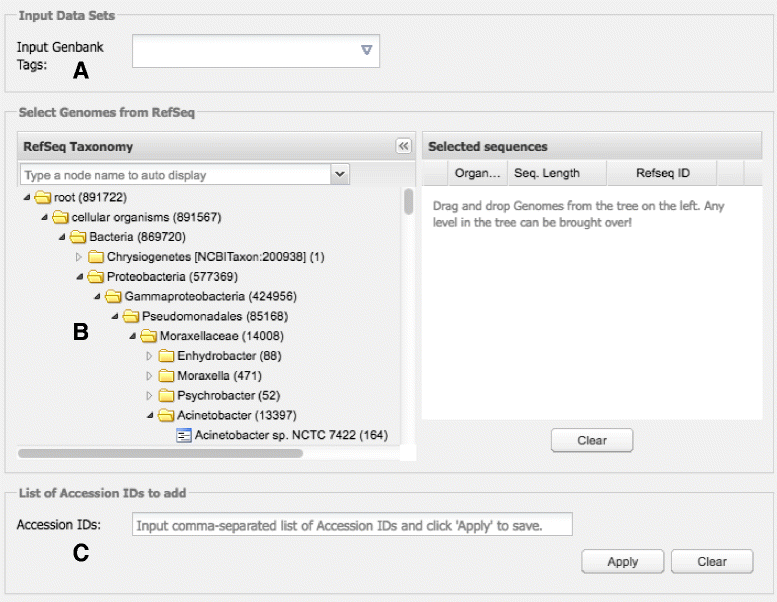
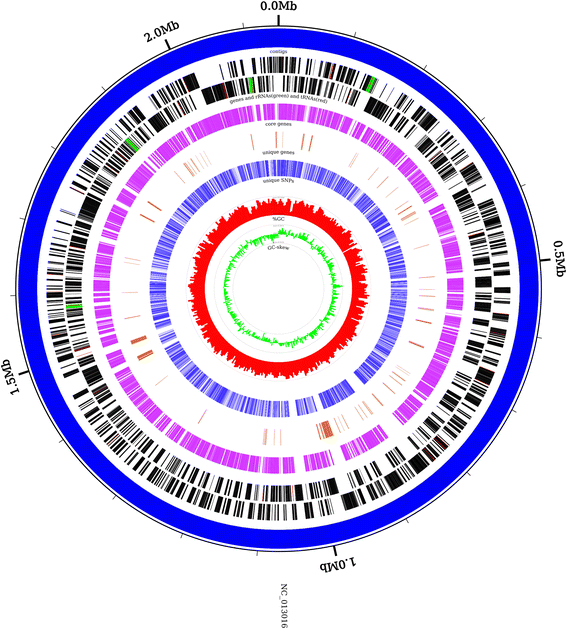
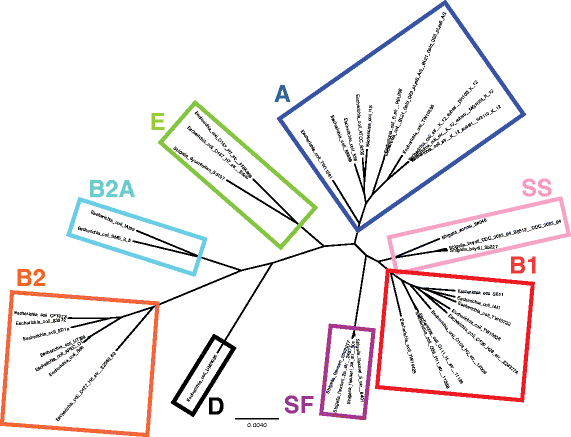
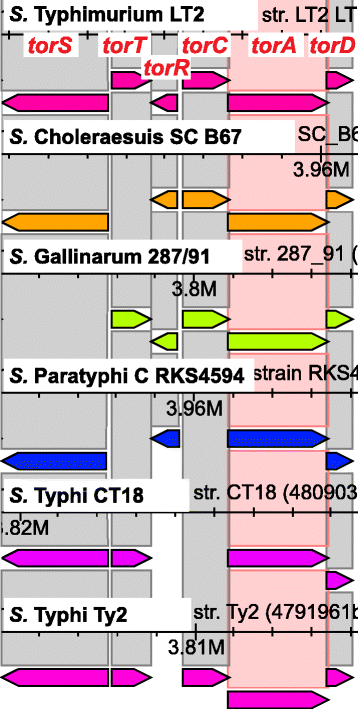
Results: CloVR-Comparative runs on annotated complete or draft genome sequences that are uploaded by the user or selected via a taxonomic tree-based user interface and downloaded from NCBI. CloVR-Comparative runs reference-free multiple whole-genome alignments to determine unique, shared and core coding sequences (CDSs) and single nucleotide polymorphisms (SNPs). Output includes short summary reports and detailed text-based results files, graphical visualizations (phylogenetic trees, circular figures), and a database file linked to the Sybil comparative genome browser. Data up- and download, pipeline configuration and monitoring, and access to Sybil are managed through CloVR-Comparative web interface. CloVR-Comparative and Sybil are distributed as part of the CloVR virtual appliance, which runs on local computers or the Amazon EC2 cloud. Representative datasets (e.g. 40 draft and complete Escherichia coli genomes) are processed in <36 h on a local desktop or at a cost of <$20 on EC2.
Conclusions: CloVR-Comparative allows anybody with Internet access to run comparative genomics projects, while eliminating the need for on-site computational resources and expertise.
Keywords: Automated analysis; Bioinformatics resource; Cloud computing; Comparative genomics; Microbial genomics; Virtual machine; Whole-genome alignment.
Figures


References
-
- Galens K, White JR, Arze C, Matalka M, Giglio MG, Team TC, Angiuoli SV, Fricke WF. Nature Preceding. 2011. CloVR-Microbe: Assembly, gene finding and functional annotation of raw sequence data from single microbial genome projects – standard operating procedure, version 1.0.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

