Structure, Genetics and Worldwide Spread of New Delhi Metallo-β-lactamase (NDM): a threat to public health
- PMID: 28449650
- PMCID: PMC5408368
- DOI: 10.1186/s12866-017-1012-8
Structure, Genetics and Worldwide Spread of New Delhi Metallo-β-lactamase (NDM): a threat to public health
Abstract
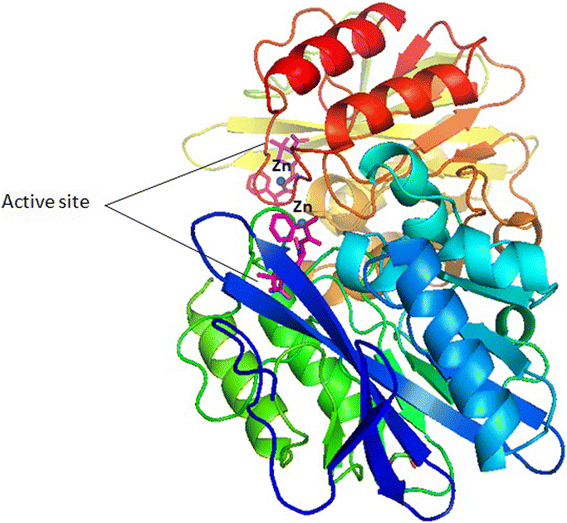
Background: The emergence of carbapenemase producing bacteria, especially New Delhi metallo-β-lactamase (NDM-1) and its variants, worldwide, has raised amajor public health concern. NDM-1 hydrolyzes a wide range of β-lactam antibiotics, including carbapenems, which are the last resort of antibiotics for the treatment of infections caused by resistant strain of bacteria.
Main body: In this review, we have discussed bla NDM-1variants, its genetic analysis including type of specific mutation, origin of country and spread among several type of bacterial species. Wide members of enterobacteriaceae, most commonly Escherichia coli, Klebsiella pneumoniae, Enterobacter cloacae, and gram-negative non-fermenters Pseudomonas spp. and Acinetobacter baumannii were found to carry these markers. Moreover, at least seventeen variants of bla NDM-type gene differing into one or two residues of amino acids at distinct positions have been reported so far among different species of bacteria from different countries. The genetic and structural studies of these variants are important to understand the mechanism of antibiotic hydrolysis as well as to design new molecules with inhibitory activity against antibiotics.
Conclusion: This review provides a comprehensive view of structural differences among NDM-1 variants, which are a driving force behind their spread across the globe.
Keywords: Antibiotic resistance; Carbapenemases; Enterobacteriaceae; New Delhi-Metallo-Beta-Lactamases.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

