Large-scale detection of drug off-targets: hypotheses for drug repurposing and understanding side-effects
- PMID: 28449705
- PMCID: PMC5408384
- DOI: 10.1186/s40360-017-0128-7
Large-scale detection of drug off-targets: hypotheses for drug repurposing and understanding side-effects
Abstract
Background: Promiscuity in molecular interactions between small-molecules, including drugs, and proteins is widespread. Such unintended interactions can be exploited to suggest drug repurposing possibilities as well as to identify potential molecular mechanisms responsible for observed side-effects.
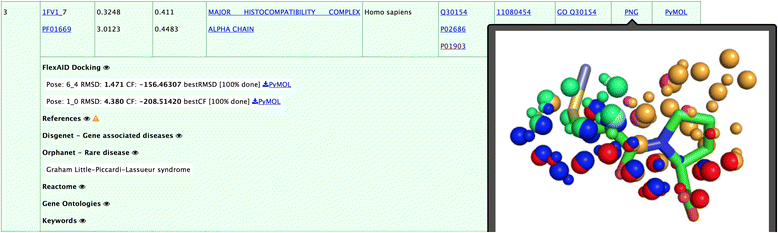
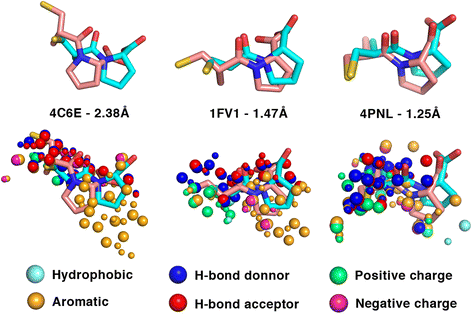
Methods: We perform a large-scale analysis to detect binding-site molecular interaction field similarities between the binding-sites of the primary target of 400 drugs against a dataset of 14082 cavities within 7895 different proteins representing a non-redundant dataset of all proteins with known structure. Statistically-significant cases with high levels of similarities represent potential cases where the drugs that bind the original target may in principle bind the suggested off-target. Such cases are further analysed with docking simulations to verify if indeed the drug could, in principle, bind the off-target. Diverse sources of data are integrated to associated potential cross-reactivity targets with side-effects.
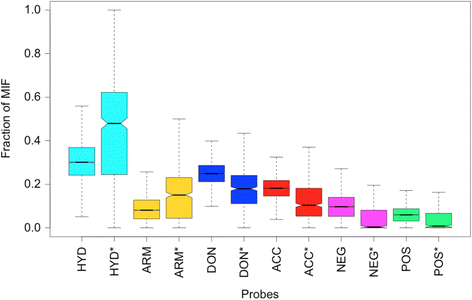
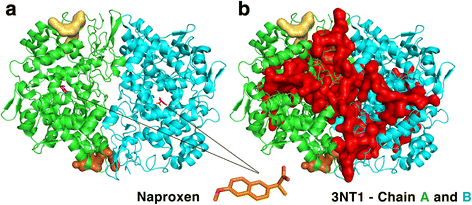
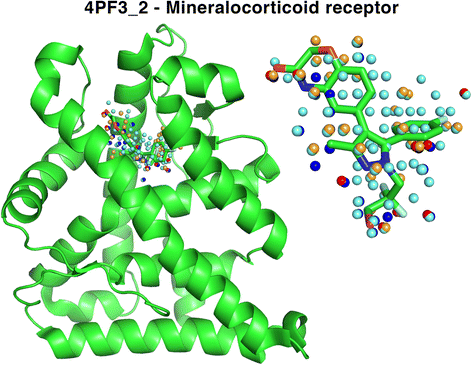
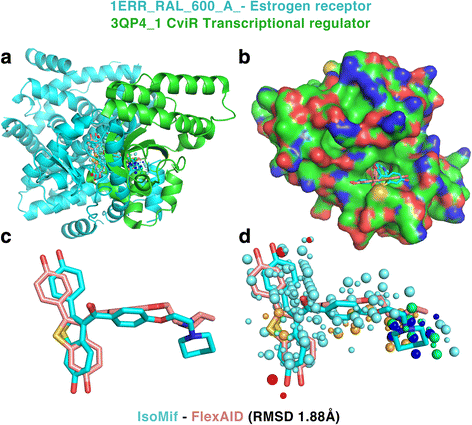
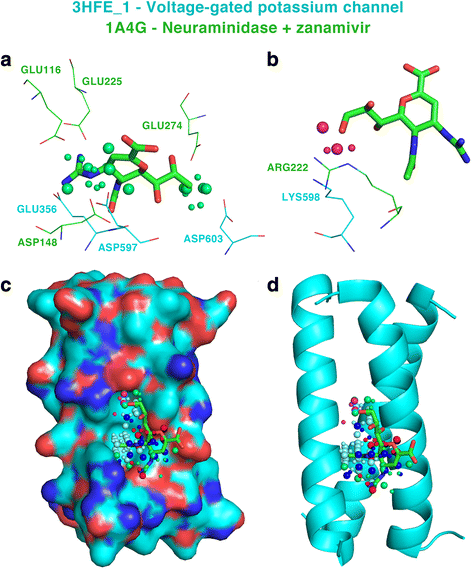
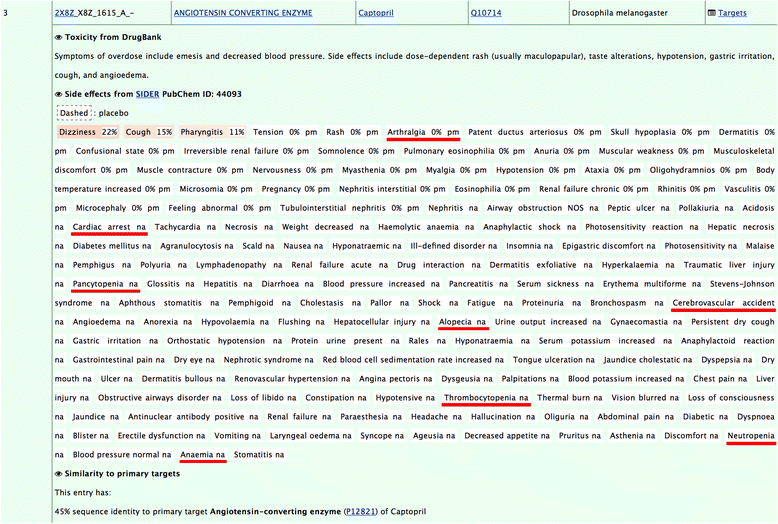
Results: We observe that promiscuous binding-sites tend to display higher levels of hydrophobic and aromatic similarities. Focusing on the most statistically significant similarities (Z-score ≥ 3.0) and corroborating docking results (RMSD < 2.0 Å), we find 2923 cases involving 140 unique drugs and 1216 unique potential cross-reactivity protein targets. We highlight a few cases with a potential for drug repurposing (acetazolamide as a chorismate pyruvate lyase inhibitor, raloxifene as a bacterial quorum sensing inhibitor) as well as to explain the side-effects of zanamivir and captopril. A web-interface permits to explore the detected similarities for each of the 400 binding-sites of the primary drug targets and visualise them for the most statistically significant cases.
Conclusions: The detection of molecular interaction field similarities provide the opportunity to suggest drug repurposing opportunities as well as to identify potential molecular mechanisms responsible for side-effects. All methods utilized are freely available and can be readily applied to new query binding-sites. All data is freely available and represents an invaluable source to identify further candidates for repurposing and suggest potential mechanisms responsible for side-effects.
Keywords: Binding-site similarities; Cross-reactivity; Drug repurposing; Large-scale analysis; Molecularinteraction field similarities; Promiscuity; Side-effects.
Figures


References
-
- Peters J-U. Polypharmacology – Foe or Friend? J Med Chem. Am Chem Soc. 2013;56:8955–71. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

