Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naïve Parkinson's disease patients
- PMID: 28449715
- PMCID: PMC5408370
- DOI: 10.1186/s13073-017-0428-y
Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naïve Parkinson's disease patients
Erratum in
-
Erratum to: Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naïve Parkinson's disease patients.Genome Med. 2017 Jun 29;9(1):61. doi: 10.1186/s13073-017-0451-z. Genome Med. 2017. PMID: 28662719 Free PMC article. No abstract available.
Abstract
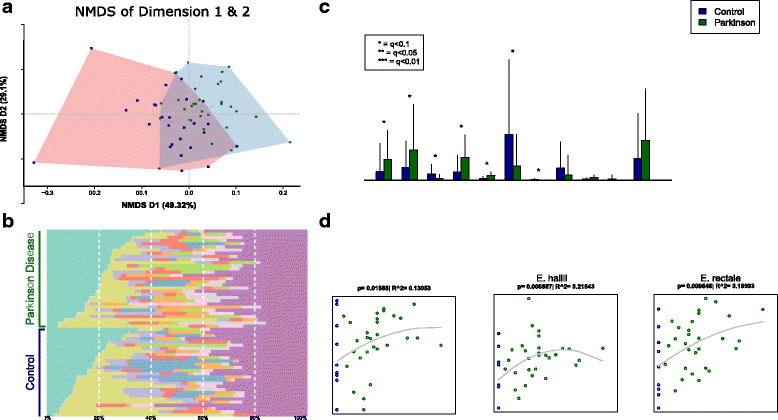
Background: Parkinson's disease (PD) presently is conceptualized as a protein aggregation disease in which pathology involves both the enteric and the central nervous system, possibly spreading from one to another via the vagus nerves. As gastrointestinal dysfunction often precedes or parallels motor symptoms, the enteric system with its vast diversity of microorganisms may be involved in PD pathogenesis. Alterations in the enteric microbial taxonomic level of L-DOPA-naïve PD patients might also serve as a biomarker.
Methods: We performed metagenomic shotgun analyses and compared the fecal microbiomes of 31 early stage, L-DOPA-naïve PD patients to 28 age-matched controls.
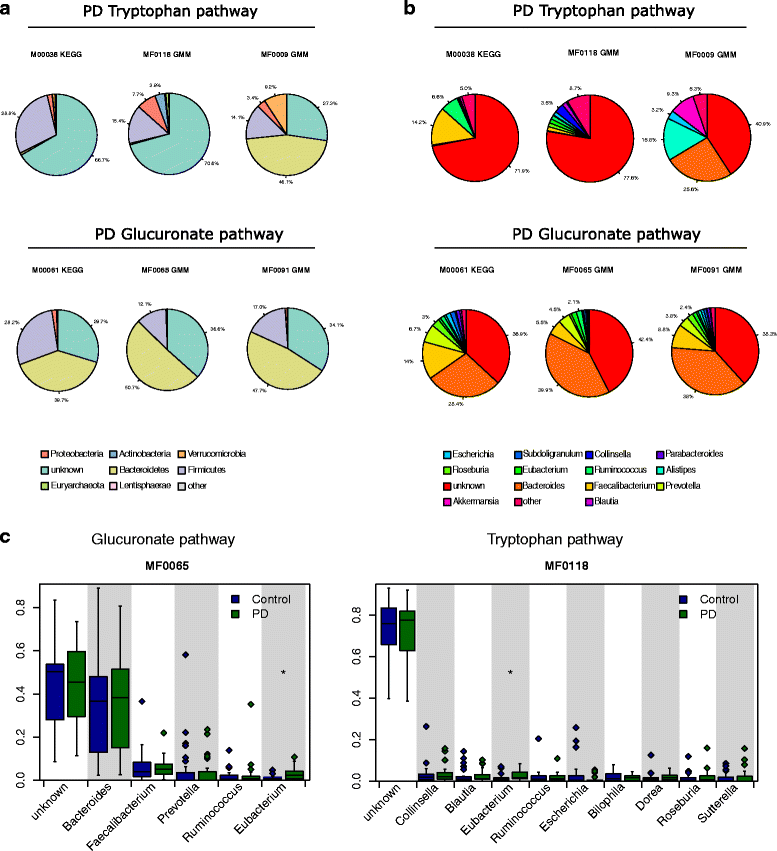
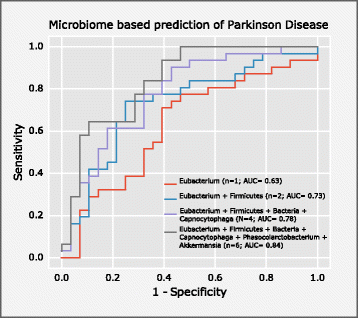
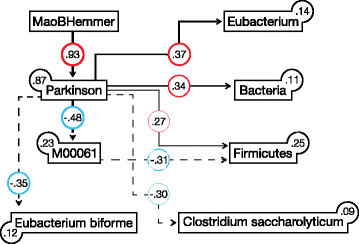
Results: We found increased Verrucomicrobiaceae (Akkermansia muciniphila) and unclassified Firmicutes, whereas Prevotellaceae (Prevotella copri) and Erysipelotrichaceae (Eubacterium biforme) were markedly lowered in PD samples. The observed differences could reliably separate PD from control with a ROC-AUC of 0.84. Functional analyses of the metagenomes revealed differences in microbiota metabolism in PD involving the ẞ-glucuronate and tryptophan metabolism. While the abundances of prophages and plasmids did not differ between PD and controls, total virus abundance was decreased in PD participants. Based on our analyses, the intake of either a MAO inhibitor, amantadine, or a dopamine agonist (which in summary relates to 90% of PD patients) had no overall influence on taxa abundance or microbial functions.
Conclusions: Our data revealed differences of colonic microbiota and of microbiota metabolism between PD patients and controls at an unprecedented detail not achievable through 16S sequencing. The findings point to a yet unappreciated aspect of PD, possibly involving the intestinal barrier function and immune function in PD patients. The influence of the parkinsonian medication should be further investigated in the future in larger cohorts.
Keywords: Archaea; Bacteria; Enteric nervous system; Gut-brain axis; Microbiome; Parkinson; Viruses.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

