DNA methylation: an epigenetic mark of cellular memory
- PMID: 28450738
- PMCID: PMC6130213
- DOI: 10.1038/emm.2017.10
DNA methylation: an epigenetic mark of cellular memory
Abstract
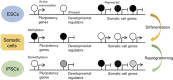
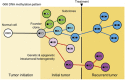
DNA methylation is a stable epigenetic mark that can be inherited through multiple cell divisions. During development and cell differentiation, DNA methylation is dynamic, but some DNA methylation patterns may be retained as a form of epigenetic memory. DNA methylation profiles can be useful for the lineage classification and quality control of stem cells such as embryonic stem cells, induced pluripotent cells and mesenchymal stem cells. During cancer initiation and progression, genome-wide and gene-specific DNA methylation changes occur as a consequence of mutated or deregulated chromatin regulators. Early aberrant DNA methylation states occurring during transformation appear to be retained during tumor evolution. Similarly, DNA methylation differences among different regions of a tumor reflect the history of cancer cells and their response to the tumor microenvironment. Therefore, DNA methylation can be a useful molecular marker for cancer diagnosis and drug treatment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell 2007; 128: 669–681. - PubMed
-
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 2002; 16: 6–21. - PubMed
-
- Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington’s canal. Nat Rev Mol Cell Biol 2009; 10: 526–537. - PubMed
-
- Shipony Z, Mukamel Z, Cohen NM, Landan G, Chomsky E, Zeliger SR et al. Dynamic and static maintenance of epigenetic memory in pluripotent and somatic cells. Nature 2014; 513: 115–119. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

