Pan-Genomic Analysis Permits Differentiation of Virulent and Non-virulent Strains of Xanthomonas arboricola That Cohabit Prunus spp. and Elucidate Bacterial Virulence Factors
- PMID: 28450852
- PMCID: PMC5389983
- DOI: 10.3389/fmicb.2017.00573
Pan-Genomic Analysis Permits Differentiation of Virulent and Non-virulent Strains of Xanthomonas arboricola That Cohabit Prunus spp. and Elucidate Bacterial Virulence Factors
Abstract
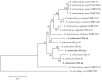
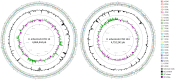
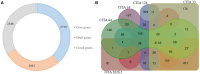
Xanthomonas arboricola is a plant-associated bacterial species that causes diseases on several plant hosts. One of the most virulent pathovars within this species is X. arboricola pv. pruni (Xap), the causal agent of bacterial spot disease of stone fruit trees and almond. Recently, a non-virulent Xap-look-a-like strain isolated from Prunus was characterized and its genome compared to pathogenic strains of Xap, revealing differences in the profile of virulence factors, such as the genes related to the type III secretion system (T3SS) and type III effectors (T3Es). The existence of this atypical strain arouses several questions associated with the abundance, the pathogenicity, and the evolutionary context of X. arboricola on Prunus hosts. After an initial characterization of a collection of Xanthomonas strains isolated from Prunus bacterial spot outbreaks in Spain during the past decade, six Xap-look-a-like strains, that did not clustered with the pathogenic strains of Xap according to a multi locus sequence analysis, were identified. Pathogenicity of these strains was analyzed and the genome sequences of two Xap-look-a-like strains, CITA 14 and CITA 124, non-virulent to Prunus spp., were obtained and compared to those available genomes of X. arboricola associated with this host plant. Differences were found among the genomes of the virulent and the Prunus non-virulent strains in several characters related to the pathogenesis process. Additionally, a pan-genomic analysis that included the available genomes of X. arboricola, revealed that the atypical strains associated with Prunus were related to a group of non-virulent or low virulent strains isolated from a wide host range. The repertoire of the genes related to T3SS and T3Es varied among the strains of this cluster and those strains related to the most virulent pathovars of the species, corylina, juglandis, and pruni. This variability provides information about the potential evolutionary process associated to the acquisition of pathogenicity and host specificity in X. arboricola. Finally, based in the genomic differences observed between the virulent and the non-virulent strains isolated from Prunus, a sensitive and specific real-time PCR protocol was designed to detect and identify Xap strains. This method avoids miss-identifications due to atypical strains of X. arboricola that can cohabit Prunus.
Keywords: almond; bacterial spot disease; comparative genomics; stone fruit trees.
Figures


References
-
- Anonymous (2000). Council directive 2000/29/EC of 8 May 2000 on protective measures against the introduction into the community of organism harmful to plants or plant products and against their spread within the community. Off. J. Eur. Commun. L 169, 1–112.
-
- Ah-You N., Gagnevin L., Chiroleu F., Jouen E., Neto J. R., Pruvost O. (2007). Pathological variations within Xanthomonas campestris pv. mangiferaindicae support its separation into three distinct pathovars that can be distinguished by amplified fragment length polymorphism. Phytopathology 97, 1568–1577. 10.1094/PHYTO-97-12-1568 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

