Comparative Analysis of Ralstonia solanacearum Methylomes
- PMID: 28450872
- PMCID: PMC5390034
- DOI: 10.3389/fpls.2017.00504
Comparative Analysis of Ralstonia solanacearum Methylomes
Abstract
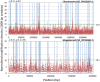
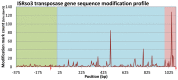
Ralstonia solanacearum is an important soil-borne plant pathogen with broad geographical distribution and the ability to cause wilt disease in many agriculturally important crops. Genome sequencing of multiple R. solanacearum strains has identified both unique and shared genetic traits influencing their evolution and ability to colonize plant hosts. Previous research has shown that DNA methylation can drive speciation and modulate virulence in bacteria, but the impact of epigenetic modifications on the diversification and pathogenesis of R. solanacearum is unknown. Sequencing of R. solanacearum strains GMI1000 and UY031 using Single Molecule Real-Time technology allowed us to perform a comparative analysis of R. solanacearum methylomes. Our analysis identified a novel methylation motif associated with a DNA methylase that is conserved in all complete Ralstonia spp. genomes and across the Burkholderiaceae, as well as a methylation motif associated to a phage-borne methylase unique to R. solanacearum UY031. Comparative analysis of the conserved methylation motif revealed that it is most prevalent in gene promoter regions, where it displays a high degree of conservation detectable through phylogenetic footprinting. Analysis of hyper- and hypo-methylated loci identified several genes involved in global and virulence regulatory functions whose expression may be modulated by DNA methylation. Analysis of genome-wide modification patterns identified a significant correlation between DNA modification and transposase genes in R. solanacearum UY031, driven by the presence of a high copy number of ISrso3 insertion sequences in this genome and pointing to a novel mechanism for regulation of transposition. These results set a firm foundation for experimental investigations into the role of DNA methylation in R. solanacearum evolution and its adaptation to different plants.
Keywords: Ralstonia; comparative genomics; epigenomics; genome; methylome; nucleotide modification; transcriptional regulation; transposon.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

