Insertion and deletion polymorphisms of the ancient AluS family in the human genome
- PMID: 28450901
- PMCID: PMC5402677
- DOI: 10.1186/s13100-017-0089-9
Insertion and deletion polymorphisms of the ancient AluS family in the human genome
Abstract
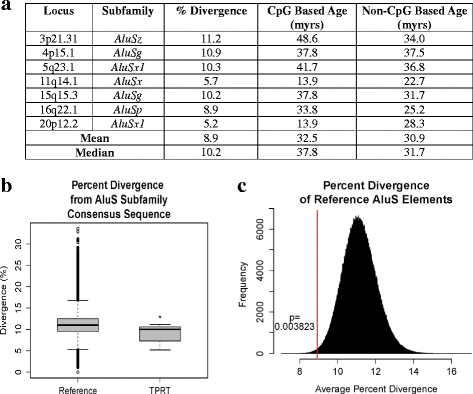
Background: Polymorphic Alu elements account for 17% of structural variants in the human genome. The majority of these belong to the youngest AluY subfamilies, and most structural variant discovery efforts have focused on identifying Alu polymorphisms from these currently retrotranspositionally active subfamilies. In this report we analyze polymorphisms from the evolutionarily older AluS subfamily, whose peak activity was tens of millions of years ago. We annotate the AluS polymorphisms, assess their likely mechanism of origin, and evaluate their contribution to structural variation in the human genome.
Results: Of 52 previously reported polymorphic AluS elements ascertained for this study, 48 were confirmed to belong to the AluS subfamily using high stringency subfamily classification criteria. Of these, the majority (77%, 37/48) appear to be deletion polymorphisms. Two polymorphic AluS elements (4%) have features of non-classical Alu insertions and one polymorphic AluS element (2%) likely inserted by a mechanism involving internal priming. Seven AluS polymorphisms (15%) appear to have arisen by the classical target-primed reverse transcription (TPRT) retrotransposition mechanism. These seven TPRT products are 3' intact with 3' poly-A tails, and are flanked by target site duplications; L1 ORF2p endonuclease cleavage sites were also observed, providing additional evidence that these are L1 ORF2p endonuclease-mediated TPRT insertions. Further sequence analysis showed strong conservation of both the RNA polymerase III promoter and SRP9/14 binding sites, important for mediating transcription and interaction with retrotransposition machinery, respectively. This conservation of functional features implies that some of these are fairly recent insertions since they have not diverged significantly from their respective retrotranspositionally competent source elements.
Conclusions: Of the polymorphic AluS elements evaluated in this report, 15% (7/48) have features consistent with TPRT-mediated insertion, thus suggesting that some AluS elements have been more active recently than previously thought, or that fixation of AluS insertion alleles remains incomplete. These data expand the potential significance of polymorphic AluS elements in contributing to structural variation in the human genome. Future discovery efforts focusing on polymorphic AluS elements are likely to identify more such polymorphisms, and approaches tailored to identify deletion alleles may be warranted.
Keywords: Alu; AluS; Mobile element; Mobilome; Polymorphism; Retrotransposon; SINE; Structural variation.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

