Specific inhibition of CK2α from an anchor outside the active site
- PMID: 28451126
- PMCID: PMC5355960
- DOI: 10.1039/c6sc02335e
Specific inhibition of CK2α from an anchor outside the active site
Abstract
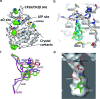
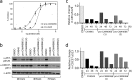
The development of selective inhibitors of protein kinases is challenging because of the significant conservation of the ATP binding site. Here, we describe a new mechanism by which the protein kinase CK2α can be selectively inhibited using features outside the ATP site. We have identified a new binding site for small molecules on CK2α adjacent to the ATP site and behind the αD loop, termed the αD pocket. An elaborated fragment anchored in this site has been linked with a low affinity fragment binding in the ATP site, creating a novel and selective inhibitor (CAM4066) that binds CK2α with a Kd of 320 nM and shows significantly improved selectivity compared to other CK2α inhibitors. CAM4066 shows target engagement in several cell lines and similar potency to clinical trial candidate CX4945. Our data demonstrate that targeting a poorly conserved, cryptic pocket allows inhibition of CK2α via a novel mechanism, enabling the development of a new generation of selective CK2α inhibitors.
Figures


References
-
- Vulpetti A., Bosotti R. Farmaco. 2004;59:759–765. - PubMed
-
- Kitagawa D., Yokota K., Gouda M., Narumi Y., Ohmoto H., Nishiwaki E., Akita K., Kirii Y. Genes Cells. 2013;18:110–122. - PubMed
-
- Davis M. I., Hunt J. P., Herrgard S., Ciceri P., Wodicka L. M., Pallares G., Hocker M., Treiber D. K., Zarrinkar P. P. Nat. Biotechnol. 2011;29:1046–1051. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

