Fluorogenic sensor platform for the histone code using receptors from dynamic combinatorial libraries
- PMID: 28451282
- PMCID: PMC5390788
- DOI: 10.1039/c6sc03003c
Fluorogenic sensor platform for the histone code using receptors from dynamic combinatorial libraries
Abstract
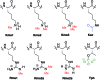
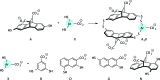
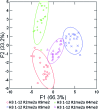
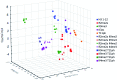
Post-translational modifications (PTMs) on histone tails act in diverse combinations in the 'histone code' to control gene expression, with dysregulation observed in a variety of diseases. However, detection and sensing methods are limited, expensive, and/or low-throughput, including MS and antibody based detection. We found that by combining four synthetic receptors developed by dynamic combinatorial chemistry (DCC) in an indicator displacement system, we are able to create a pattern-based sensor platform that can discriminate single PTMs such as methylation and acetylation on a representative histone peptide with 100% accuracy as well as peptides bearing both dimethyl and trimethyl lysine in the presence of arginine methylation, which has not previously been demonstrated, and can even correctly distinguish the position of lysine methylation individually or in the presence of other PTMs. To extend this approach, a full panel of thirteen analytes containing different combinations of PTMs were classified with 96 ± 1% overall accuracy in a 50% left-out analysis, demonstrating the robustness and versatility of the sensor array. Finally, the sensor platform was also used to demonstrate proof of concept for enzymatic assays by analysing the mock reaction of a threonine kinase, successfully identifying analytes representative of substrate conversion both with and without neighboring PTMs. This work provides a rapid platform for the analysis of peptides bearing complex modifications and highlights the utility of receptors discovered though DCC that display variations in binding affinity and selectivity.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

