DNA-barcoded labeling probes for highly multiplexed Exchange-PAINT imaging
- PMID: 28451377
- PMCID: PMC5380918
- DOI: 10.1039/c6sc05420j
DNA-barcoded labeling probes for highly multiplexed Exchange-PAINT imaging
Abstract
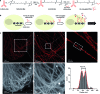
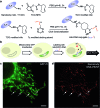
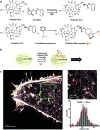
Recent advances in super-resolution fluorescence imaging allow researchers to overcome the classical diffraction limit of light, and are already starting to make an impact in biology. However, a key challenge for traditional super-resolution methods is their limited multiplexing capability, which prevents a systematic understanding of multi-protein interactions on the nanoscale. Exchange-PAINT, a recently developed DNA-based multiplexing approach, in theory facilitates spectrally-unlimited multiplexing by sequentially imaging target molecules using orthogonal dye-labeled 'imager' strands. While this approach holds great promise for the bioimaging community, its widespread application has been hampered by the availability of DNA-conjugated ligands for protein labeling. Herein, we report a universal approach for the creation of DNA-barcoded labeling probes for highly multiplexed Exchange-PAINT imaging, using a variety of affinity reagents such as primary and secondary antibodies, nanobodies, and small molecule binders. Furthermore, we extend the availability of orthogonal imager strands for Exchange-PAINT to over 50 and assay their orthogonality in a novel DNA origami-based crosstalk assay. Using our optimized conjugation and labeling strategies, we demonstrate nine-color super-resolution imaging in situ in fixed cells.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

