Enhanced degradation of polyhydroxyalkanoates (PHAs) by newly isolated Burkholderia cepacia DP1 with high depolymerase activity
- PMID: 28452023
- PMCID: PMC5428109
- DOI: 10.1007/s13205-017-0716-7
Enhanced degradation of polyhydroxyalkanoates (PHAs) by newly isolated Burkholderia cepacia DP1 with high depolymerase activity
Abstract
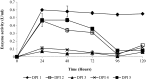
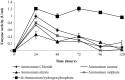
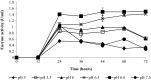
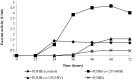
The contribution of microbial depolymerase has received much attention because of its potential in biopolymer degradation. In this study, the P(3HB) depolymerase enzyme of a newly isolated Burkholderia cepacia DP1 from soil in Penang, Malaysia, was optimized using response surface methodology (RSM). The factors affecting P(3HB) depolymerase enzyme production were studied using one-variable-at-a-time approach prior to optimization. Preliminary experiments revealed that the concentration of nitrogen source, concentration of carbon source, initial pH and incubation time were among the main factors influencing the enzyme productivity. An increase of 9.4 folds in enzyme production with an activity of 5.66 U/mL was obtained using optimal medium containing 0.028% N of di-ammonium hydrogen phosphate and 0.31% P(3HB-co-21%4HB) as carbon source at the initial pH of 6.8 for 38 h of incubation. Moreover, the RSM model showed great similarity between predicted and actual enzyme production indicating a successful model validation. This study warrants the ability of P(3HB) degradation by B. cepacia DP1 in producing higher enzyme activity as compared to other P(3HB) degraders being reported. Interestingly, the production of P(3HB) depolymerase was rarely reported within genus Burkholderia. Therefore, this is considered to be a new discovery in the field of P(3HB) depolymerase production.
Keywords: Biodegradable; Extracellular P(3HB) depolymerase enzyme; P(3HB-co-4HB); Poly(3-hydroxybutyrate) [P(3HB)]; Polyhydroxyalkanoates (PHAs); Response surface methodology (RSM).
Conflict of interest statement
The authors declare that they have no conflict of interest in the publication.
Figures





References
-
- Boyandin AN, Prudnikova SV, Karpov VA, Ivonin VN, Ðô͂ NL, Nguyê͂n TH, Gitelson II. Microbial degradation of polyhydroxyalkanoates in tropical soils. Int Biodeterior Biodegrad. 2012;83:77–84. doi: 10.1016/j.ibiod.2013.04.014. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

