Understanding structure-function relationships of the human neuronal acetylcholine receptor: insights from the first crystal structures of neuronal subunits
- PMID: 28452148
- PMCID: PMC5980119
- DOI: 10.1111/bph.13838
Understanding structure-function relationships of the human neuronal acetylcholine receptor: insights from the first crystal structures of neuronal subunits
Abstract
Nicotinic ACh receptors (nAChRs) are the best studied members of the superfamily of pentameric ligand-gated ion channels (pLGICs). Neuronal nAChRs regulate neuronal excitability and neurotransmitter release in the nervous system and form either homo- or hetero-pentameric complexes with various combinations of the 11 neuronal nAChR subunits (α2-7, α9, α10 and β2-4) known to exist in humans. In addition to their wide distribution in the nervous system, neuronal nAChRs have been also found in immune cells and many peripheral tissues. These nAChRs are important drug targets for neurological and neuropsychiatric diseases (e.g. Alzheimer's, schizophrenia) and substance addiction (e.g. nicotine), as well as in a variety of diseases such as chronic pain, auditory disorders and some cancers. To decipher the functional mechanisms of human nAChRs and develop efficient and specific therapeutic drugs, elucidation of their high-resolution structures is needed. Recent studies, including the X-ray crystal structures of the near-intact α4β2 nAChR and of the ligand-binding domains of the α9 and α2 subunits, have advanced our knowledge on the detailed structure of the ligand-binding sites formed between the same and different subunits and revealed many other functionally important interactions. The aim of this review is to highlight some of the structural and functional findings of these studies and to compare them with recent breakthrough findings on other pLGIC members and earlier data from their homologous ACh-binding proteins.
Linked articles: This article is part of a themed section on Nicotinic Acetylcholine Receptors. To view the other articles in this section visit http://onlinelibrary.wiley.com/doi/10.1111/bph.v175.11/issuetoc.
© 2017 The British Pharmacological Society.
Figures
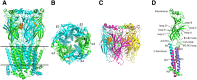

Similar articles
-
Proteins and chemical chaperones involved in neuronal nicotinic receptor expression and function: an update.Br J Pharmacol. 2018 Jun;175(11):1869-1879. doi: 10.1111/bph.13777. Epub 2017 Apr 19. Br J Pharmacol. 2018. PMID: 28294298 Free PMC article. Review.
-
α9- and α7-containing receptors mediate the pro-proliferative effects of nicotine in the A549 adenocarcinoma cell line.Br J Pharmacol. 2018 Jun;175(11):1957-1972. doi: 10.1111/bph.13954. Epub 2017 Sep 8. Br J Pharmacol. 2018. PMID: 28726253 Free PMC article.
-
Orthosteric and allosteric potentiation of heteromeric neuronal nicotinic acetylcholine receptors.Br J Pharmacol. 2018 Jun;175(11):1805-1821. doi: 10.1111/bph.13745. Epub 2017 Mar 20. Br J Pharmacol. 2018. PMID: 28199738 Free PMC article. Review.
-
Nicotinic acetylcholine receptors: from structure to brain function.Rev Physiol Biochem Pharmacol. 2003;147:1-46. doi: 10.1007/s10254-003-0005-1. Epub 2003 Mar 20. Rev Physiol Biochem Pharmacol. 2003. PMID: 12783266 Review.
-
Recent advances in understanding the structure of nicotinic acetylcholine receptors.IUBMB Life. 2009 Apr;61(4):407-23. doi: 10.1002/iub.170. IUBMB Life. 2009. PMID: 19319967 Review.
Cited by
-
A triad of residues is functionally transferrable between 5-HT3 serotonin receptors and nicotinic acetylcholine receptors.J Biol Chem. 2018 Feb 23;293(8):2903-2914. doi: 10.1074/jbc.M117.810432. Epub 2018 Jan 3. J Biol Chem. 2018. PMID: 29298898 Free PMC article.
-
The α9α10 nicotinic acetylcholine receptor: a compelling drug target for hearing loss?Expert Opin Ther Targets. 2022 Mar;26(3):291-302. doi: 10.1080/14728222.2022.2047931. Epub 2022 Mar 7. Expert Opin Ther Targets. 2022. PMID: 35225139 Free PMC article. Review.
-
Potency- and Selectivity-Enhancing Mutations of Conotoxins for Nicotinic Acetylcholine Receptors Can Be Predicted Using Accurate Free-Energy Calculations.Mar Drugs. 2021 Jun 25;19(7):367. doi: 10.3390/md19070367. Mar Drugs. 2021. PMID: 34202022 Free PMC article.
-
Neurotransmitters: promising immune modulators in the tumor microenvironment.Front Immunol. 2023 May 5;14:1118637. doi: 10.3389/fimmu.2023.1118637. eCollection 2023. Front Immunol. 2023. PMID: 37215113 Free PMC article. Review.
-
Mutations of the nACh Receptor M4 Helix Reveal Different Phenotypes in Different Expression Systems: Could Lipids be Responsible?Front Physiol. 2022 May 4;13:850782. doi: 10.3389/fphys.2022.850782. eCollection 2022. Front Physiol. 2022. PMID: 35600303 Free PMC article.
References
-
- Ahring PK, Olsen JA, Nielsen EO, Peters D, Pedersen MHF, Rohde LA et al. (2015). Engineered α4β2 nicotinic acetylcholine receptors as models for measuring agonist binding and effect at the orthosteric low‐affinity α4‐α4 interface. Neuropharmacology 92: 135–145. - PubMed
-
- Arias HR (2010). Positive and negative modulation of nicotinic receptors. Adv Protein Chem Struct Biol 80: 153–203. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

