HMGA1 amplifies Wnt signalling and expands the intestinal stem cell compartment and Paneth cell niche
- PMID: 28452345
- PMCID: PMC5414379
- DOI: 10.1038/ncomms15008
HMGA1 amplifies Wnt signalling and expands the intestinal stem cell compartment and Paneth cell niche
Abstract
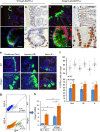
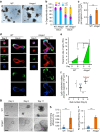
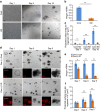
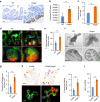
High-mobility group A1 (Hmga1) chromatin remodelling proteins are enriched in intestinal stem cells (ISCs), although their function in this setting was unknown. Prior studies showed that Hmga1 drives hyperproliferation, aberrant crypt formation and polyposis in transgenic mice. Here we demonstrate that Hmga1 amplifies Wnt/β-catenin signalling to enhance self-renewal and expand the ISC compartment. Hmga1 upregulates genes encoding both Wnt agonist receptors and downstream Wnt effectors. Hmga1 also helps to 'build' an ISC niche by expanding the Paneth cell compartment and directly inducing Sox9, which is required for Paneth cell differentiation. In human intestine, HMGA1 and SOX9 are positively correlated, and both become upregulated in colorectal cancer. Our results define a unique role for Hmga1 in intestinal homeostasis by maintaining the stem cell pool and fostering terminal differentiation to establish an epithelial stem cell niche. This work also suggests that deregulated Hmga1 perturbs this equilibrium during intestinal carcinogenesis.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

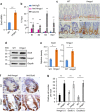
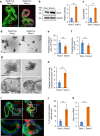

References
-
- Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell 154, 274–284 (2013). - PubMed
-
- Koo B. K. et al.. Controlled gene expression in primary Lgr5 organoid cultures. Nat. Methods 9, 81–83 (2011). - PubMed
-
- Koo B. K., Sasselli V. & Clevers H. Retroviral gene expression control in primary organoid cultures. Curr. Protoc. Stem Cell Biol. 27, 5A.6.1-5A.6.8 (2013). - PubMed
-
- Sato T. & Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science 340, 1190–1194 (2013). - PubMed
-
- Koo B. K. & Clevers H. Stem cells marked by the R-spondin receptor LGR5. Gastroenterology 147, 289–302 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

