Establishment of expanded and streamlined pipeline of PITCh knock-in - a web-based design tool for MMEJ-mediated gene knock-in, PITCh designer, and the variations of PITCh, PITCh-TG and PITCh-KIKO
- PMID: 28453368
- PMCID: PMC5470537
- DOI: 10.1080/21655979.2017.1313645
Establishment of expanded and streamlined pipeline of PITCh knock-in - a web-based design tool for MMEJ-mediated gene knock-in, PITCh designer, and the variations of PITCh, PITCh-TG and PITCh-KIKO
Abstract
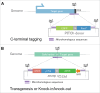
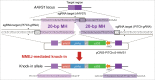
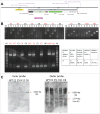
The emerging genome editing technology has enabled the creation of gene knock-in cells easily, efficiently, and rapidly, which has dramatically accelerated research in the field of mammalian functional genomics, including in humans. We recently developed a microhomology-mediated end-joining-based gene knock-in method, termed the PITCh system, and presented various examples of its application. Since the PITCh system only requires very short microhomologies (up to 40 bp) and single-guide RNA target sites on the donor vector, the targeting construct can be rapidly prepared compared with the conventional targeting vector for homologous recombination-based knock-in. Here, we established a streamlined pipeline to design and perform PITCh knock-in to further expand the availability of this method by creating web-based design software, PITCh designer ( http://www.mls.sci.hiroshima-u.ac.jp/smg/PITChdesigner/index.html ), as well as presenting an experimental example of versatile gene cassette knock-in. PITCh designer can automatically design not only the appropriate microhomologies but also the primers to construct locus-specific donor vectors for PITCh knock-in. By using our newly established pipeline, a reporter cell line for monitoring endogenous gene expression, and transgenesis (TG) or knock-in/knockout (KIKO) cell line can be produced systematically. Using these new variations of PITCh, an exogenous promoter-driven gene cassette expressing fluorescent protein gene and drug resistance gene can be integrated into a safe harbor or a specific gene locus to create transgenic reporter cells (PITCh-TG) or knockout cells with reporter knock-in (PITCh-KIKO), respectively.
Keywords: CRISPR–Cas9; gene knock-in; genome engineering; microhomology-mediated end-joining (MMEJ); web tool.
Figures
References
-
- Sakuma T, Woltjen K. Nuclease-mediated genome editing: At the front-line of functional genomics technology. Dev Growth Differ 2014; 56(1):2-13; PMID:24387662; https://doi.org/ 10.1111/dgd.12111 - DOI - PubMed
-
- Sakuma T, Nakade S, Sakane Y, Suzuki KT, Yamamoto T. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat Protoc 2016; 11(1):118-33; PMID:26678082; https://doi.org/ 10.1038/nprot.2015.140 - DOI - PubMed
-
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al.. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol 2009; 27(9):851-7; PMID:19680244; https://doi.org/ 10.1038/nbt.1562 - DOI - PMC - PubMed
-
- Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, Hatanaka F, Yamamoto M, Araoka T, Li Z, et al.. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 2016; 540(7631):144-9; PMID:27851729; https://doi.org/ 10.1038/nature20565 - DOI - PMC - PubMed
-
- Nakade S, Tsubota T, Sakane Y, Kume S, Sakamoto N, Obara M, Daimon T, Sezutsu H, Yamamoto T, Sakuma T, et al.. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat Commun 2014; 5:5560; PMID:25410609; https://doi.org/ 10.1038/ncomms6560 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
