Pioneering topological methods for network-based drug-target prediction by exploiting a brain-network self-organization theory
- PMID: 28453640
- PMCID: PMC6291778
- DOI: 10.1093/bib/bbx041
Pioneering topological methods for network-based drug-target prediction by exploiting a brain-network self-organization theory
Abstract
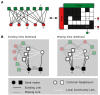
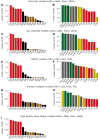
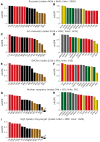
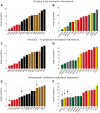
The bipartite network representation of the drug-target interactions (DTIs) in a biosystem enhances understanding of the drugs' multifaceted action modes, suggests therapeutic switching for approved drugs and unveils possible side effects. As experimental testing of DTIs is costly and time-consuming, computational predictors are of great aid. Here, for the first time, state-of-the-art DTI supervised predictors custom-made in network biology were compared-using standard and innovative validation frameworks-with unsupervised pure topological-based models designed for general-purpose link prediction in bipartite networks. Surprisingly, our results show that the bipartite topology alone, if adequately exploited by means of the recently proposed local-community-paradigm (LCP) theory-initially detected in brain-network topological self-organization and afterwards generalized to any complex network-is able to suggest highly reliable predictions, with comparable performance with the state-of-the-art-supervised methods that exploit additional (non-topological, for instance biochemical) DTI knowledge. Furthermore, a detailed analysis of the novel predictions revealed that each class of methods prioritizes distinct true interactions; hence, combining methodologies based on diverse principles represents a promising strategy to improve drug-target discovery. To conclude, this study promotes the power of bio-inspired computing, demonstrating that simple unsupervised rules inspired by principles of topological self-organization and adaptiveness arising during learning in living intelligent systems (like the brain) can efficiently equal perform complicated algorithms based on advanced, supervised and knowledge-based engineering.
Figures





References
-
- Paul SM, Mytelka DS, Dunwiddie CT, et al.How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat Rev Drug Discov 2010;9(3):203–14. - PubMed
-
- Munos B. Lessons from 60 years of pharmaceutical innovation. Nat Rev Drug Discov 2009;8(12):959–68. - PubMed
-
- O’Connor KA, Roth BL.. Finding new tricks for old drugs: an efficient route for public-sector drug discovery. Nat Rev Drug Discov 2005;4(12):1005–14. - PubMed
-
- Yamanishi Y. Supervised Bipartite Graph Inference. NIPS, 2008, 1841–1848. MIT press, Cambridge, USA.
-
- Bleakley K, Biau G, Vert J-P.. Supervised reconstruction of biological networks with local models. Bioinformatics 2007;23(13):i57–65. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

