Progranulin functions as a cathepsin D chaperone to stimulate axonal outgrowth in vivo
- PMID: 28453791
- PMCID: PMC5886064
- DOI: 10.1093/hmg/ddx162
Progranulin functions as a cathepsin D chaperone to stimulate axonal outgrowth in vivo
Abstract
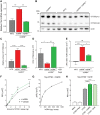
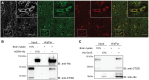
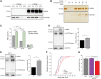
Loss of function mutations in progranulin (GRN) cause frontotemporal dementia, but how GRN haploinsufficiency causes neuronal dysfunction remains unclear. We previously showed that GRN is neurotrophic in vitro. Here, we used an in vivo axonal outgrowth system and observed a delayed recovery in GRN-/- mice after facial nerve injury. This deficit was rescued by reintroduction of human GRN and relied on its C-terminus and on neuronal GRN production. Transcriptome analysis of the facial motor nucleus post injury identified cathepsin D (CTSD) as the most upregulated gene. In aged GRN-/- cortices, CTSD was also upregulated, but the relative CTSD activity was reduced and improved upon exogenous GRN addition. Moreover, GRN and its C-terminal granulin domain granulinE (GrnE) both stimulated the proteolytic activity of CTSD in vitro. Pull-down experiments confirmed a direct interaction between GRN and CTSD. This interaction was also observed with GrnE and stabilized the CTSD enzyme at different temperatures. Investigating the importance of this interaction for axonal regeneration in vivo we found that, although individually tolerated, a combined reduction of GRN and CTSD synergistically reduced axonal outgrowth. Our data links the neurotrophic effect of GRN and GrnE with a lysosomal chaperone function on CTSD to maintain its proteolytic capacity.
© The Author 2017. Published by Oxford University Press.
Figures
References
-
- Bateman A., Bennett H.P. (1998) Granulins: the structure and function of an emerging family of growth factors. J. Endocrinol., 158, 145–151. - PubMed
-
- Zhu J., Nathan C., Jin W., Sim D., Ashcroft G.S., Wahl S.M., Lacomis L., Erdjument-Bromage H., Tempst P., Wright C.D.. et al. (2002) Conversion of proepithelin to epithelins: roles of SLPI and elastase in host defense and wound repair. Cell, 111, 867–878. - PubMed
-
- De Muynck L., Van Damme P. (2011) Cellular effects of progranulin in health and disease. J. Mol. Neurosci., 45, 549–560. - PubMed
-
- Petkau T.L., Leavitt B.R. (2014) Progranulin in neurodegenerative disease. Trends Neurosci., 37, 388–398. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

