Longitudinal genomic surveillance of Plasmodium falciparum malaria parasites reveals complex genomic architecture of emerging artemisinin resistance
- PMID: 28454557
- PMCID: PMC5410087
- DOI: 10.1186/s13059-017-1204-4
Longitudinal genomic surveillance of Plasmodium falciparum malaria parasites reveals complex genomic architecture of emerging artemisinin resistance
Abstract
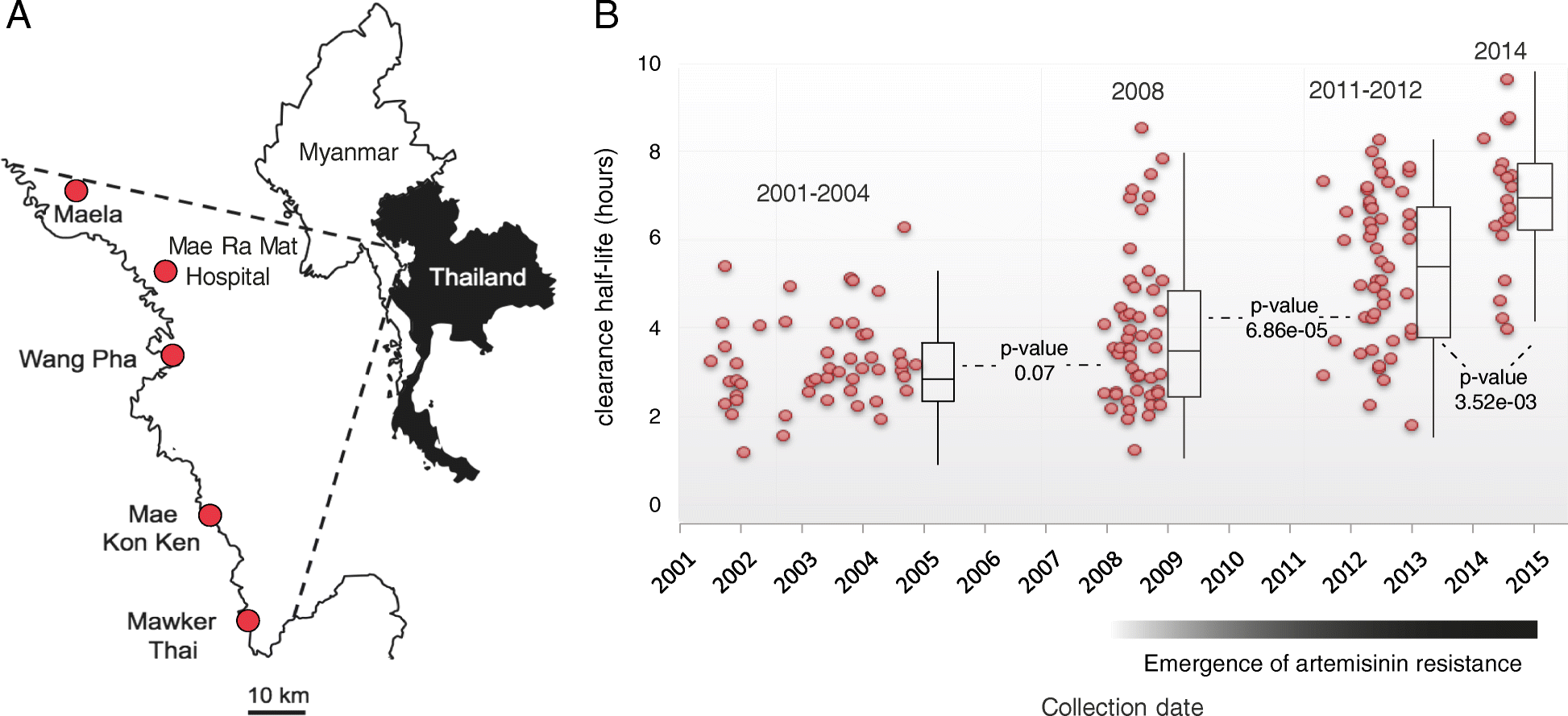
Background: Artemisinin-based combination therapies are the first line of treatment for Plasmodium falciparum infections worldwide, but artemisinin resistance has risen rapidly in Southeast Asia over the past decade. Mutations in the kelch13 gene have been implicated in this resistance. We used longitudinal genomic surveillance to detect signals in kelch13 and other loci that contribute to artemisinin or partner drug resistance. We retrospectively sequenced the genomes of 194 P. falciparum isolates from five sites in Northwest Thailand, over the period of a rapid increase in the emergence of artemisinin resistance (2001-2014).
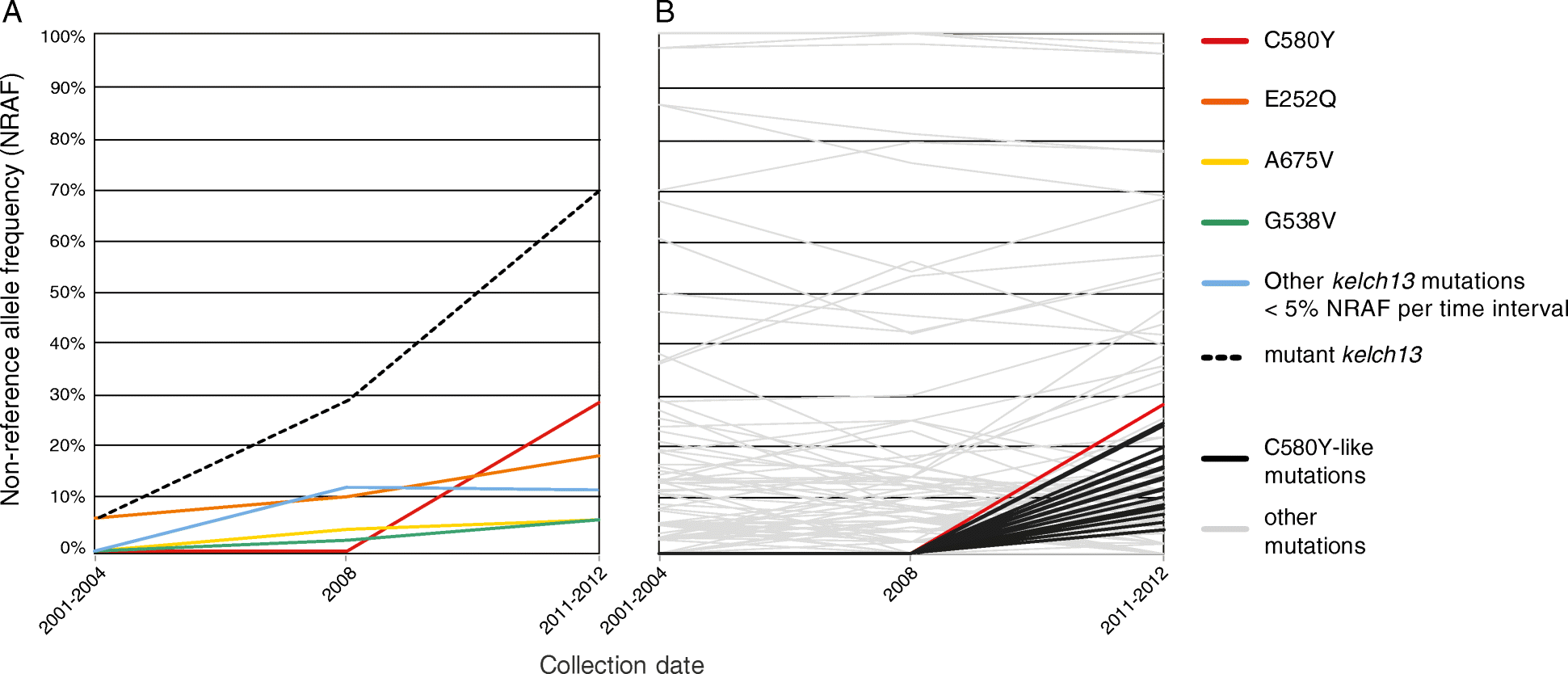
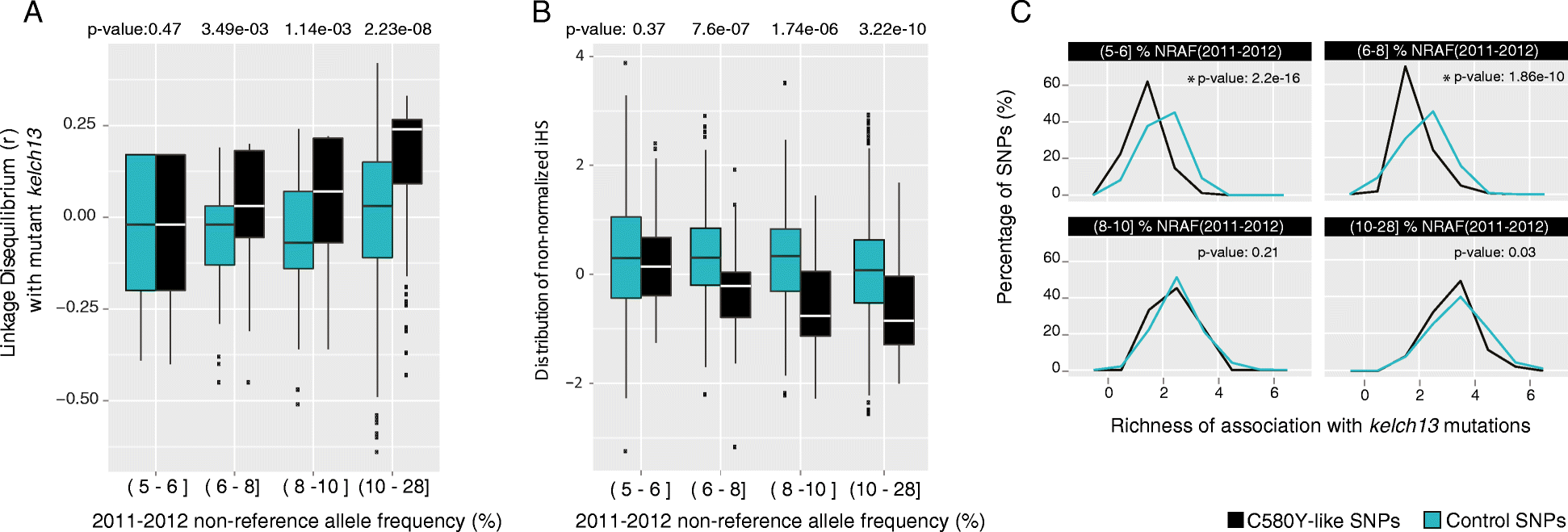
Results: We evaluate statistical metrics for temporal change in the frequency of individual SNPs, assuming that SNPs associated with resistance increase in frequency over this period. After Kelch13-C580Y, the strongest temporal change is seen at a SNP in phosphatidylinositol 4-kinase, which is involved in a pathway recently implicated in artemisinin resistance. Furthermore, other loci exhibit strong temporal signatures which warrant further investigation for involvement in artemisinin resistance evolution. Through genome-wide association analysis we identify a variant in a kelch domain-containing gene on chromosome 10 that may epistatically modulate artemisinin resistance.
Conclusions: This analysis demonstrates the potential of a longitudinal genomic surveillance approach to detect resistance-associated gene loci to improve our mechanistic understanding of how resistance develops. Evidence for additional genomic regions outside of the kelch13 locus associated with artemisinin-resistant parasites may yield new molecular markers for resistance surveillance, which may be useful in efforts to reduce the emergence or spread of artemisinin resistance in African parasite populations.
Keywords: Drug resistance; Epistasis; Genomics; Malaria; Surveillance.
Figures


Comment in
-
Longitudinal study of Plasmodium pathogens identifies new loci associated with artemisinin resistance.Genome Biol. 2017 Apr 28;18(1):79. doi: 10.1186/s13059-017-1219-x. Genome Biol. 2017. PMID: 28454594 Free PMC article.
References
-
- World Health Organization . Guidelines for the treatment of malaria. 3 2015. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

